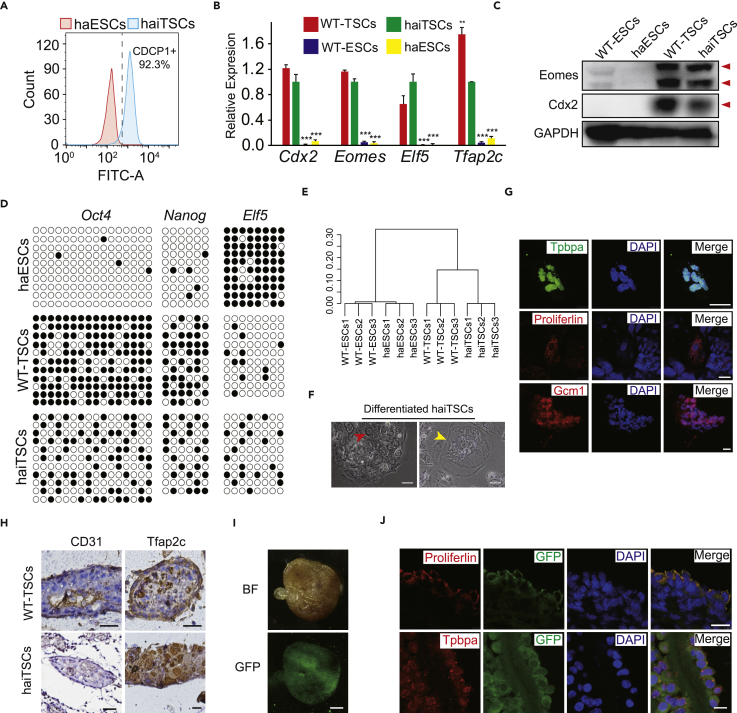
Figure 3.
Identification of haiTSCs Properties and Differentiation Potential
(A) FACS analysis of CDCP1-positive cells among established haiTSCs. haESCs are used as negative control.
(B) The expression levels of TSC-specific marker genes (Cdx2, Eomes, Elf5, and Tfap2c) in haiTSCs, WT-TSCs, haESCs, and WT-ESCs. t test, **p < 0.01, ***p < 0.001. Data are represented as mean ± SEM.
(C) Western blot analysis of Eomes and Cdx2 in WT-ESCs, haESCs, WT-TSCs, and haiTSCs. GAPDH is used as a loading control.
(D) DNA methylation status in the promoter regions of Oct4, Nanog, and Elf5. An haESCs line and a WT-TSCs line were used as controls.
(E) Global gene expression cluster analysis of transcripts in WT-ESCs, haESCs, WT-TSCs, and haiTSCs.
(F) Images of differentiated haiTSCs. Red arrow indicates syncytiotrophoblast cells, and yellow arrow indicates trophoblast giant cells. Scale bar, 100 μm.
(G) Immunofluorescence staining of three trophoblast-lineage-specific markers, Tpbpa (fluorescein isothiocyanate channel), proliferin (Tetramethylrhodamine [TRITC] channel), and GCM1 (TRITC channel), in cells differentiated from haiTSCs in vitro. DNA is stained with DAPI. Scale bar, 25 μm.
(H) Immunohistochemical (IHC) analysis of hemorrhagic lesions derived from haiTSCs. Fixed sample is IHC stained against the endothelial marker CD31 and the trophoblast marker Tfap2c.
(I) Images of BF (top) and GFP (bottom) of a chimeric placenta following blastocyst injection of haiTSCs-eGFP cells. Scale bar, 2 mm.
(J) Immunofluorescence staining of chimeric placenta following sectioning, with trophoblast-lineage-specific antibodies against proliferin and Tpbpa. Scale bar, 25 μm.
See also Figures S4 and S5.