Figure 1.
The AKT Apoptosis Model Predicts Protein Dynamics in Different Genetic Backgrounds
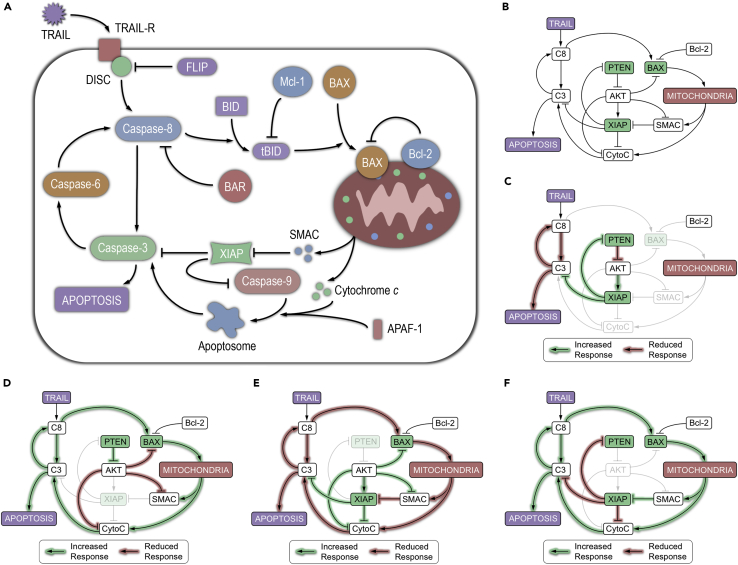
(A) Extrinsic apoptosis signaling pathway.
(B) The AKT apoptosis model (AKTM). Protein species shown in green are those for which double knockout HCT116 cell lines are used in this study. TRAIL, TNF-related apoptosis-inducing ligand; C8, active caspase-8; BAX, Bcl-2-associated X protein; Bcl-2, B-cell lymphoma 2; SMAC, second mitochondria-derived activator of caspases; CytoC, cytochrome c; XIAP, X-linked inhibitor of apoptosis protein; AKT, protein kinase B; PTEN, phosphatase and tensin homolog; C3, active caspase-3.
(C) Predicting the effects of BAX removal on the AKTM. Green lines signify a predicted increased response, and red lines signify a reduced response.
(D) Predicted effects of XIAP removal on the AKTM.
(E) Predicted effects of PTEN removal on the AKTM.
(F) Predicted effects of AKT removal on the AKTM.