An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.
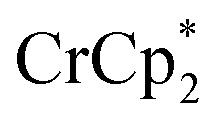 + 1 equiv of Na+), and reduction of MIL-125 with
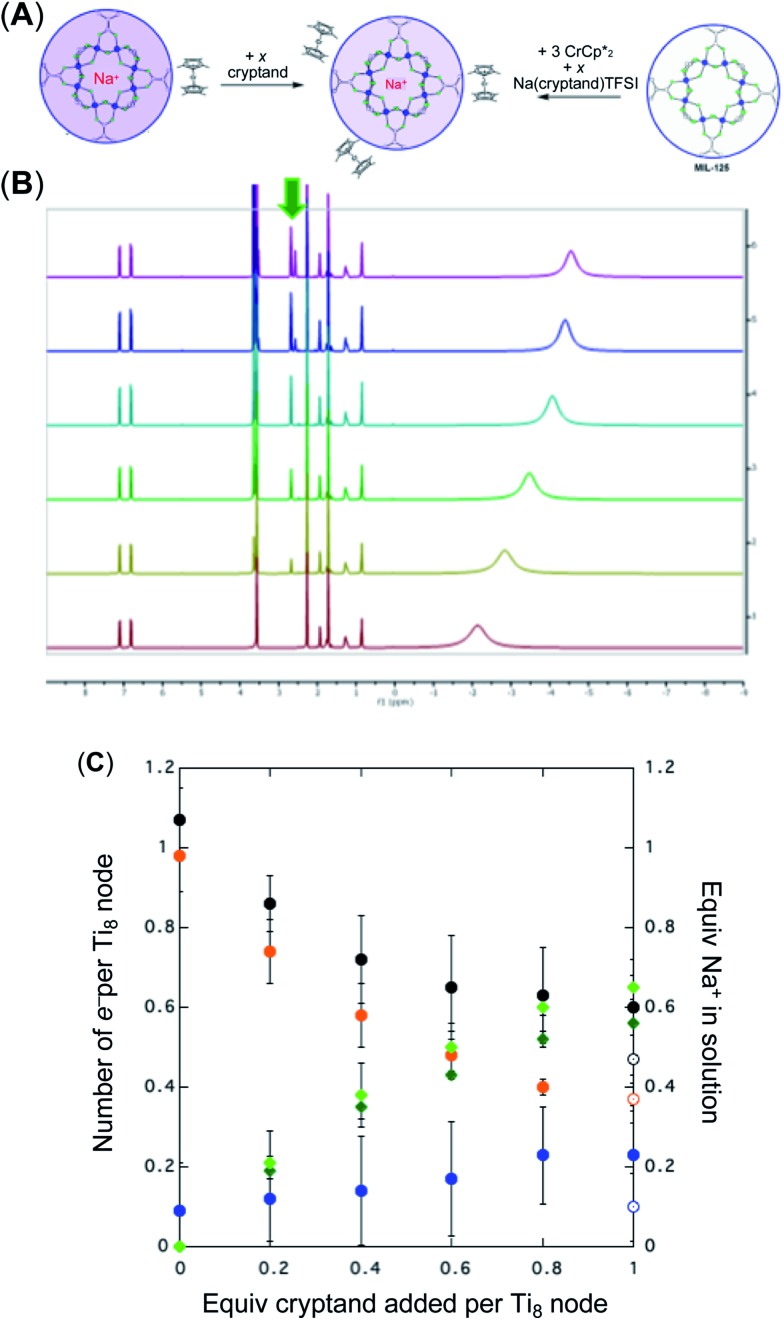
+ 1 equiv of Na+), and reduction of MIL-125 with 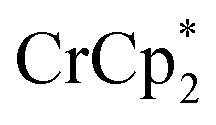 and [Na·cryptand]TFSI. (A) Schematic of this reaction. (B) 1H NMR spectra (d8-THF + CD3CN, 4 : 1 v/v) MIL-125 + 3 equiv.
and [Na·cryptand]TFSI. (A) Schematic of this reaction. (B) 1H NMR spectra (d8-THF + CD3CN, 4 : 1 v/v) MIL-125 + 3 equiv. 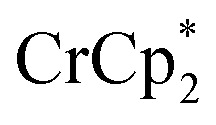 + 3 equiv. NaTFSI + 0 (red), 0.2 (yellow), 0.4 (green), 0.6 (light blue), 0.8 (dark blue), and 1.0 (purple) equiv. cryptand (per Ti8 node). The two peaks indicated by the arrow correspond to free (right) and Na-bound (left) cryptand. (C) Plot of e– in MIL-125 versus equivs of cryptand added (left axis), and equiv. of Na+ in solution (right axis). Orange circles: equivs Na+ associated with the reduced MIL-125, derived from the
+ 3 equiv. NaTFSI + 0 (red), 0.2 (yellow), 0.4 (green), 0.6 (light blue), 0.8 (dark blue), and 1.0 (purple) equiv. cryptand (per Ti8 node). The two peaks indicated by the arrow correspond to free (right) and Na-bound (left) cryptand. (C) Plot of e– in MIL-125 versus equivs of cryptand added (left axis), and equiv. of Na+ in solution (right axis). Orange circles: equivs Na+ associated with the reduced MIL-125, derived from the 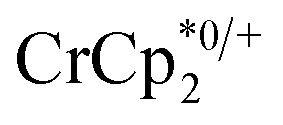 chemical shift. Blue circles: equivs
chemical shift. Blue circles: equivs 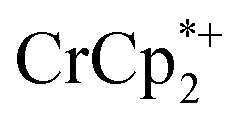 associated with the reduced MIL-125, derived from the change in integration of the
associated with the reduced MIL-125, derived from the change in integration of the 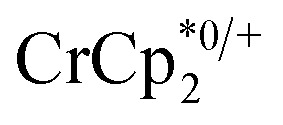 resonance. Black circles: total extent of reduction (sum of blue and orange). Light green diamonds: equiv. Na+ in solution, derived from the amount of Na+ still associated with reduced MIL-125. Dark green diamonds: equiv. of Na+ in solution, from integration of the cryptand/cryptand-sodium resonances. Open circles: number of e– in MIL-125 (per Ti8 node) for the converse reaction: addition of 3 equiv. of
resonance. Black circles: total extent of reduction (sum of blue and orange). Light green diamonds: equiv. Na+ in solution, derived from the amount of Na+ still associated with reduced MIL-125. Dark green diamonds: equiv. of Na+ in solution, from integration of the cryptand/cryptand-sodium resonances. Open circles: number of e– in MIL-125 (per Ti8 node) for the converse reaction: addition of 3 equiv. of 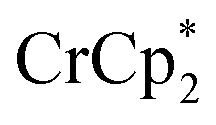 and 1 equiv. of [Na·cryptand]TFSI.
and 1 equiv. of [Na·cryptand]TFSI.