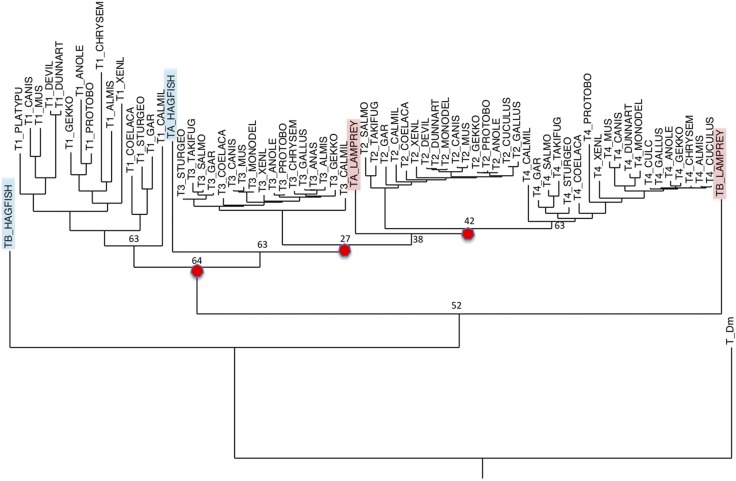
Figure 4.
A phylogenetic tree generated from vertebrate TIMP amino acid sequences. This tree was generated by using TIMP sequences, devoid of secretion signal sequences, from species representing therian and prototherian mammals, reptiles (including birds), amphibians, and jawless, cartilaginous, lobe-finned, and bony fish. The sequences were aligned by using the computer program MUSCLE (47), and the tree was generated by using RAxML (46). The tree was rooted with D. melanogaster TIMP, and sequence differences were scored by using the JTT matrix. The red circles identify the projected gene duplication events.