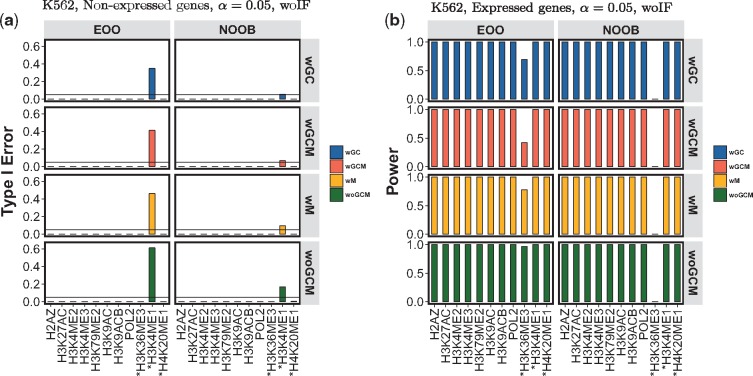
Fig. 3.
Assessment of GLANET Type-I error and power with data-driven computational experiments. Results for the two association statistics—existence of overlap (EOO) and the number of overlapping bases (NOOB)—together with GC (wGC), with Mappability (wM), with GC and Mappability (wGCM), and without GC and mappability (woGCM) null distribution generation modes are displayed. Histone marks with ambiguous activator roles are marked with *. (a, b) Type-I error and power estimated without Isochore Family (woIF) heuristic using K562, (Non-expressed Genes, CompletelyDiscard) and (Expressed Genes,Top5) results, for significance level of 0.05