Fig. 5.
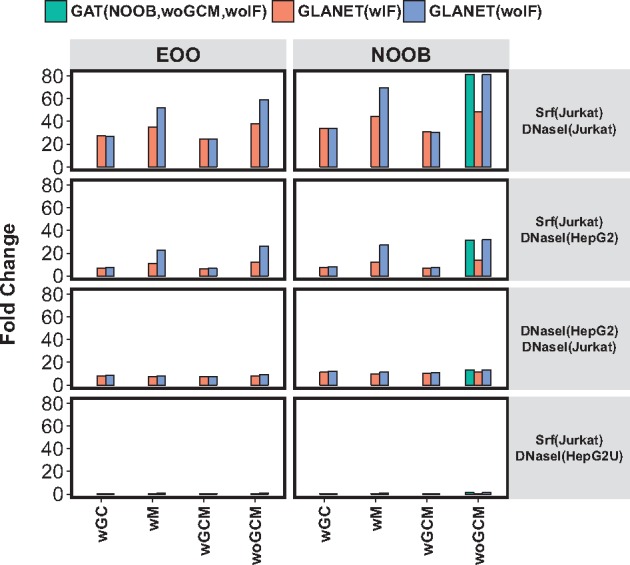
GLANET and GAT are run on four experiments ranging from high to low expected association between the compared genomic interval sets. Each row depicts an experiment where the first set is input query and the second set is a genomic element in the annotation library, e.g. experiment Srf(Jurkat) versus DNaseI(Jurkat) evaluates whether the binding regions of TF Srf in Jurkat cells are enriched for DNaseI accessible, i.e. open chromatin, regions in the same cells
