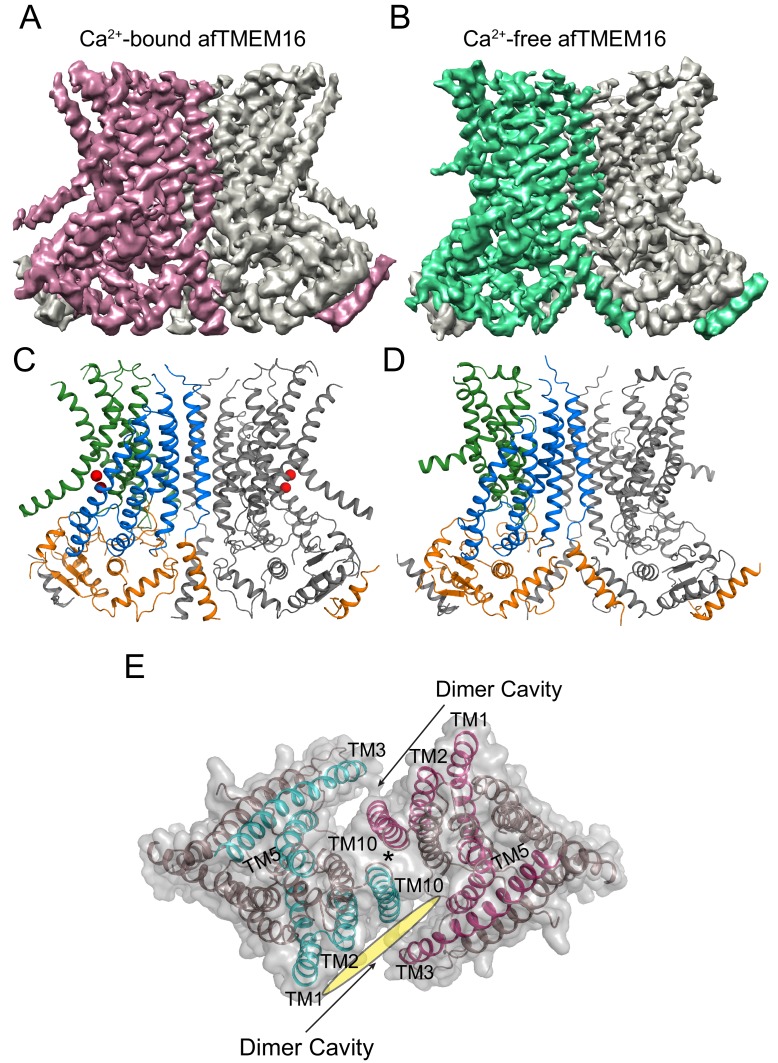
Figure 1. Structures of afTMEM16 in the presence and absence of Ca2+.
(A–B) Masked cryo EM density maps of afTMEM16 in the presence of 0.5 mM Ca2+ (A) or in Ca2+-free (B) conditions. For clarity, one monomer is shown in gray while the other is colored in red (A, 0.5 mM Ca2+) or green (B, 0 Ca2+). (C–D) atomic models of afTMEM16 reconstituted in nanodiscs in the presence of 0.5 mM Ca2+ (C) and in the absence of Ca2+ (D). For clarity one monomer is gray, in the other the cytosolic domain is orange, the permeation pathway is green and the remainder of the protein is blue. Ca2+ ions are shown as red spheres. (D) Top view of Ca2+-bound afTMEM16 shown as maroon ribbon inside its surface representation. The dimer cavities are labeled and one of the two is highlighted with a yellow oval. * denotes the protein’s twofold axis of symmetry between the two TM10’s. To illustrate the antiparallel orientation of the two dimer cavities, the cavity-lining helices (TM1, 2, 3, 5 and 10) from the two monomers are colored in cyan and red.
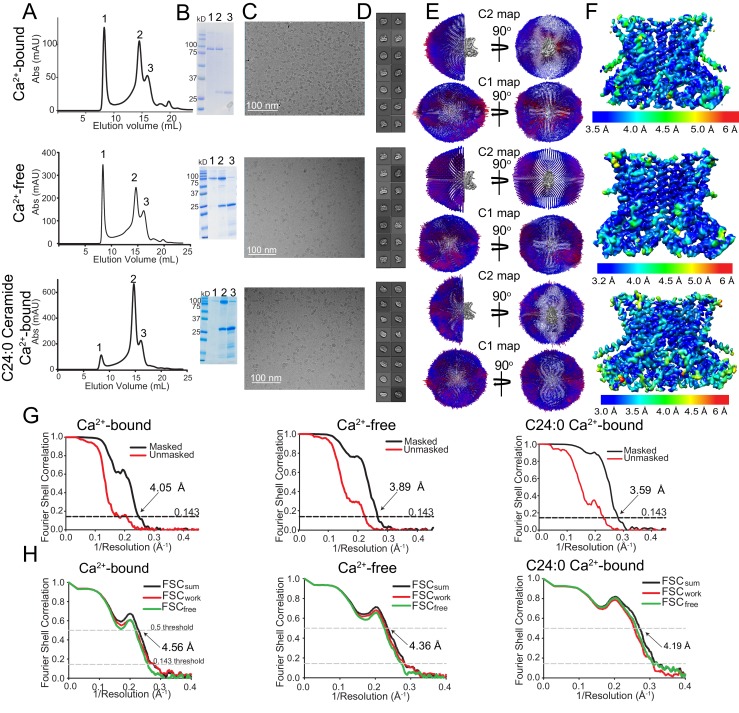
Figure 1—figure supplement 1. Cryo-EM characterization of afTMEM16/nanodisc complexes.
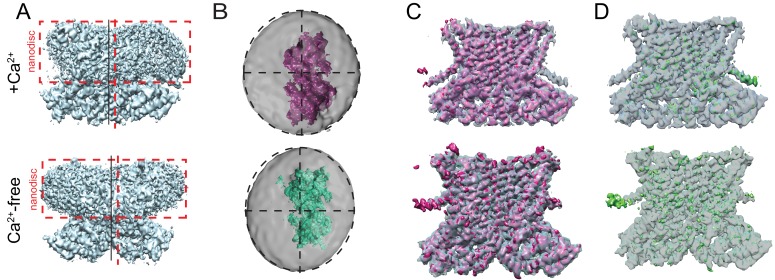
Figure 1—figure supplement 2. Asymmetry of afTMEM16-nanodisc complex.
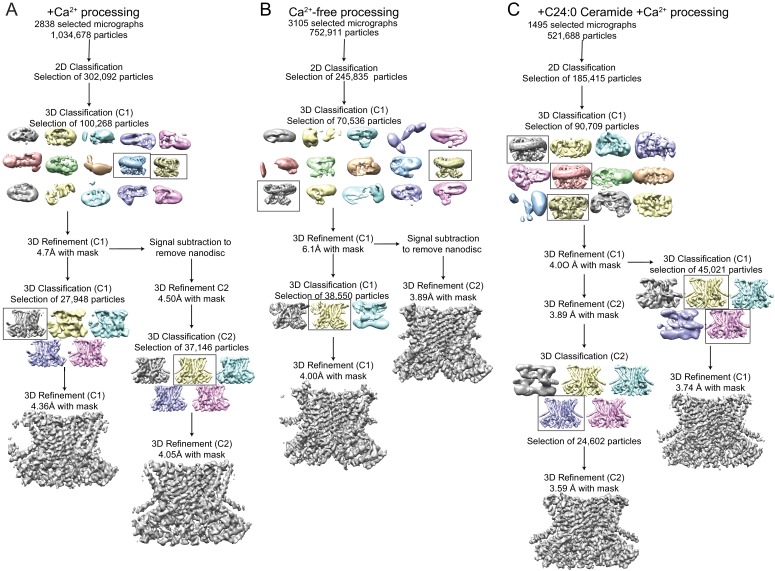
Figure 1—figure supplement 3. Cryo-EM data processing procedure for afTMEM16/nanodisc complexes.
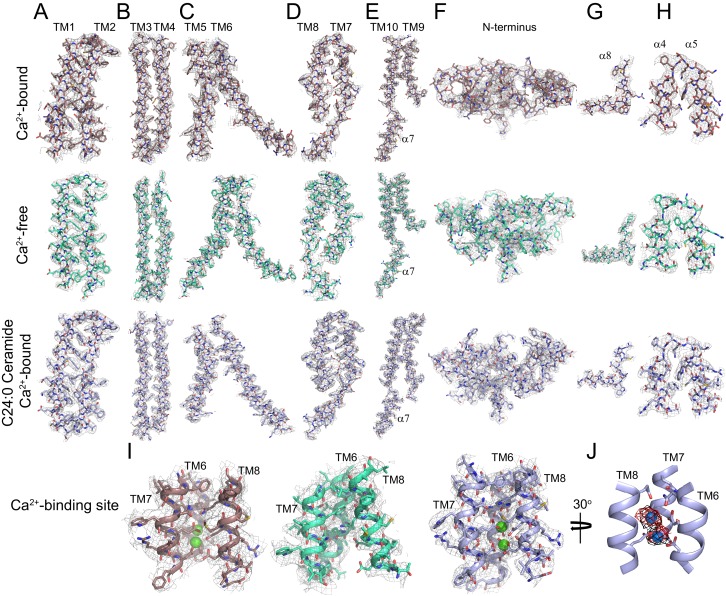
Figure 1—figure supplement 4. Representative cryo-EM density for afTMEM16 in 0.5 mM Ca2+, in 0 mM Ca2+ and in the presence of 0.5 mM Ca2+ and 5 mol% C24:0 Ceramide.
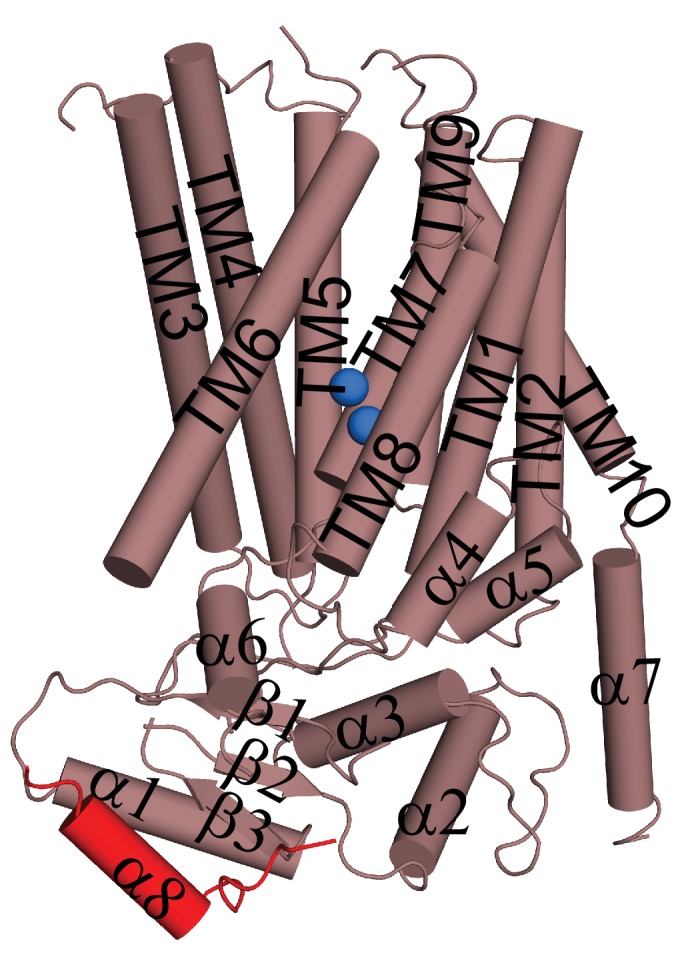
Figure 1—figure supplement 5. Helical organization of afTMEM16.