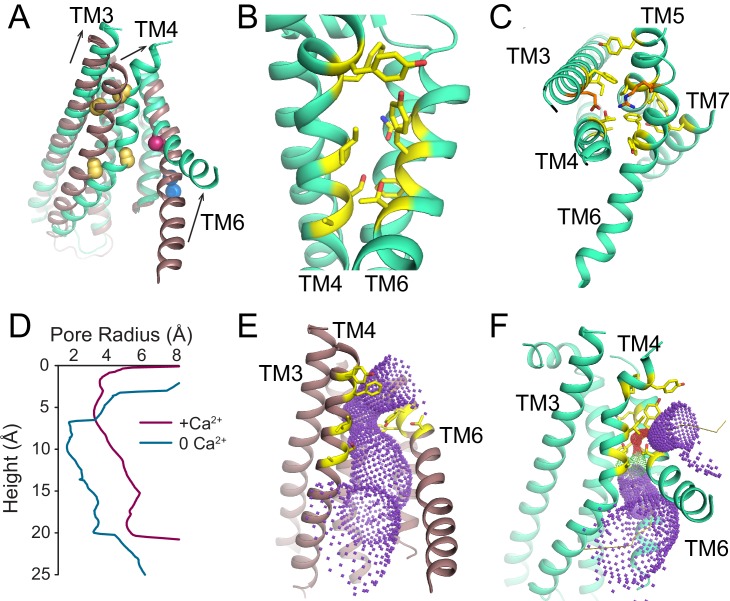
Figure 3. Ca2+-dependent rearrangements of the lipid permeation pathway.
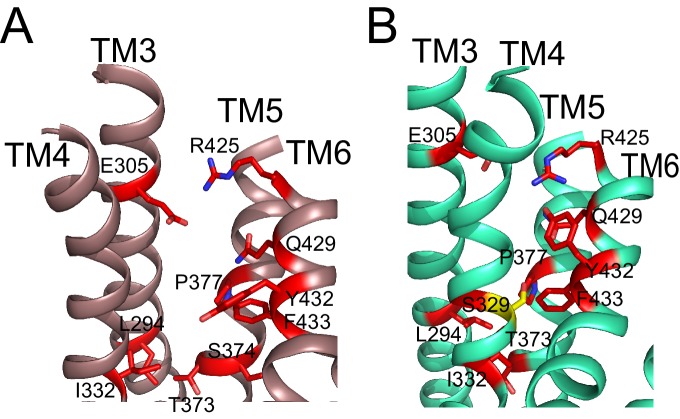
(A) Structural alignment of the lipid pathway with afTMEM16 in the presence (maroon) or absence (cyan) of 0.5 mM Ca2+. The color scheme is the same throughout the figure. Arrows indicate direction of movement from the Ca2+-free to the Ca2+-bound conformations. The lipid permeation pathway is constricted by rearrangements of TM4 around P324 and P333 (shown as yellow spheres in both structures) and TM6 at A437 (shown as a red sphere in both structures). (B) Close-up view of the closed permeation pathway, residues at the interface with the membrane are shown as yellow sticks. (C) Top view of the permeation pathway in the absence of Ca2+. Residues pointing inside the pathway are shown as yellow sticks. The interacting charged pair E305 and R425 are shown as orange sticks. (D) Diameter of the afTMEM16 lipid pathway in the presence (maroon) and absence (cyan) of Ca2+. The diameter was estimated using the HOLE program (Smart et al., 1996). (E–F) Accessibility of the lipid permeation pathway estimated using the program HOLE in the presence (E) or absence (F) of Ca2+. Purple denotes areas of diameter d > 5.5 Å, yellow areas where 5.5 < d < 2.75 Å and red areas with d < 2.75 Å.