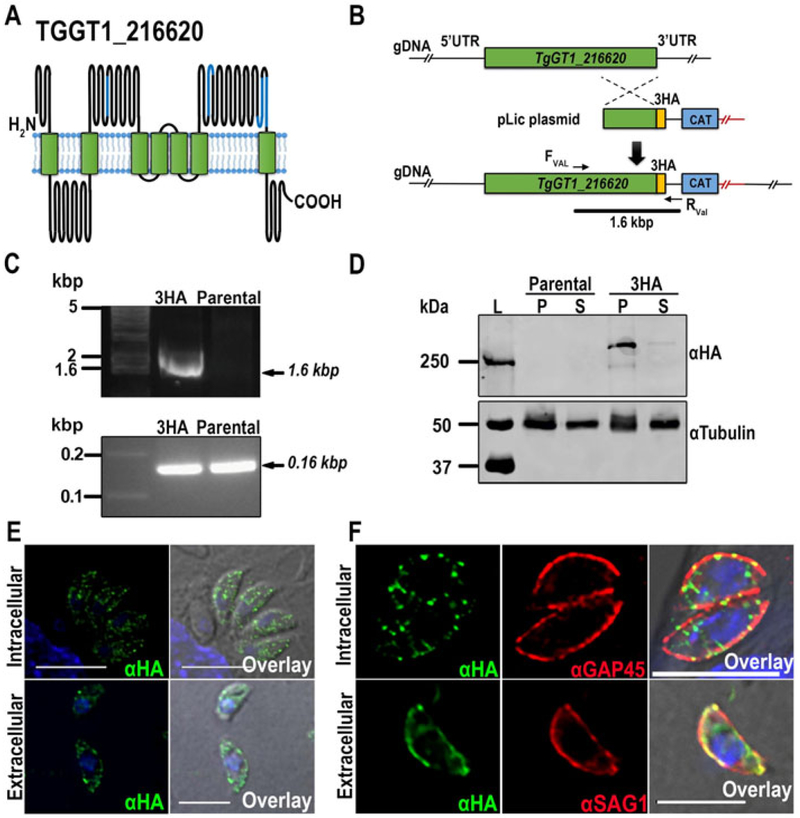
Figure 1.
Tagging and localization of TgGT1_216620. (A) Schematic model showing the predicted topology of TgGT1_216620. The EF-hand-motifs are marked in blue. The model was generated using the Protter web application (Omasits et al. 2014). (B) Scheme depicting the strategy used for the C-terminal tagging of TgGT1_216620 in the parental strain TatiΔku80. The genomic region and recombination fragment are shown in green, the C-terminal HA tag in yellow and the selection marker, chloramphenicol acetyl transferase (CAT), in blue. (C) Top panel, PCR analysis showing the correct insertion of 3HA tag (3HA) at the 3’ region of the TgGT1_216620gene. Bottom panel, PCR positive control of parental and 3HA tagged parasites. (D) Western blot analysis of total lysates from TgGT1_216620-3HA and TatIΔku80 parental cells, respectively. Top panel, two bands above ~250 kDa were detected in lysates of the tagged strain, but not in lysates of the parental strain. Bottom panel, loading controls developed with anti-tubulin. (E) Immunofluorescence analysis of intracellular and extracellular tachyzoites showing a punctuated localization of α-HA in both intracellular and extracellular parasites with a higher concentration at the plasma membrane. Scale bars = 5 μm (F) Top panel, IFAs showing co-localization of α-HA and α-GAP45 in intracellular parasites at the periphery. Bottom panel, co-localization of α-HA and α-SAG1, a plasma membrane marker, in extracellular tachyzoites (bottom). Scale bars = 5 μm