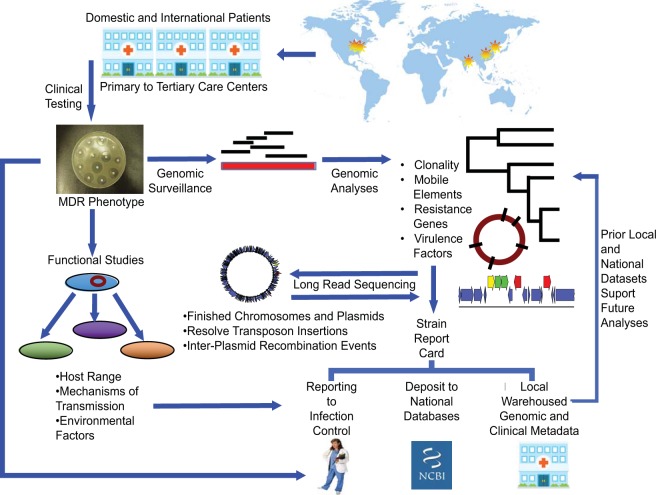
FIG 1.
In the Pathogen Genomic Surveillance Program, patients from local, regional, and international sites receive care at participating institutions. Clinically ordered testing identifies carbapenem-resistant Enterobacteriaceae, which are flagged for genome analyses with rapid-turnaround short-read sequencing to define clonal relationships, resistance genes, and mobile elements carrying resistance. Novel strains undergo long-read sequencing to refine analyses. Functional studies define the host range of mobile vectors and mechanisms of transmission. Strain report cards to local infection control teams summarize clinical, microbiologic, genomic, and functional data to evaluate outbreak potential and the landscape of strain, vector, and gene-level causes of resistance. Local warehousing of the combined information supports ongoing and future surveillance efforts. Depositing of deidentified strain antibiogram and genomic data sets into national databases supports national and international surveillance efforts.