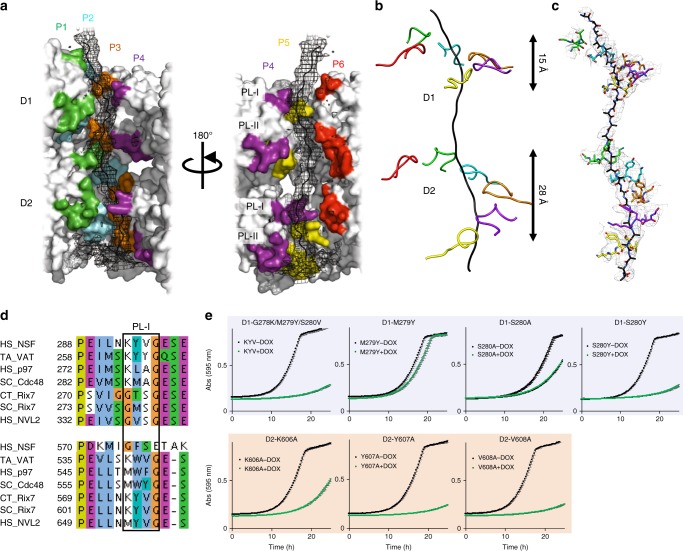
Fig. 4.
Rix7 pore loops are essential for function in vivo. a Rix7 engages a substrate in its central pore. Difference map generated from a substrate free model of Rix7 is shown in black mesh, and the modeled peptide is shown as a black cartoon. The pseudo-atomic model of Rix7 is shown as a surface representation in gray, with the pore loop residues colored by the individual protomers. The pore loop 1 and pore loop 2 are indicated as PL-I and PL-II, respectively. b Simplified view of PL-I engaging the substrate in the D1 and D2 domains. The D2 PL-I engages more of the polypeptide than the D1 PL-I. c Conserved motifs from D1 and D2 PL1 (shown as sticks) grip the polypeptide (shown as sticks). The cryo-EM map is overlaid on top. d Multiple sequence alignment of the D1 and D2 domains from several type II AAA family members, including NSF, VAT/p97/Cdc48, and Rix7/NVL2. Only the alignment surrounding PL-I (boxed) in each AAA domain is shown. Abbreviations are as follows: Homo sapiens (HS), Chaetomium thermophilum (CT), Saccharomyces cerevisiae (SC) and Thermoplasma acidophilum (TA). Alignments were done in Clustal omega69 and illustrated with JalView70. e Growth curves of S. cerevisiae tetO7-RIX7 strains transformed with plasmids encoding pore loop mutants of Rix7. Strains were grown in the absence (black) or presence (green) of doxycycline at 25 °C and the absorbance was recorded at 595 nm over a 25 h time period. Each curve is the average of three independent replicates and the error bars mark the standard deviation between replicates. Source data are provided as a source data file