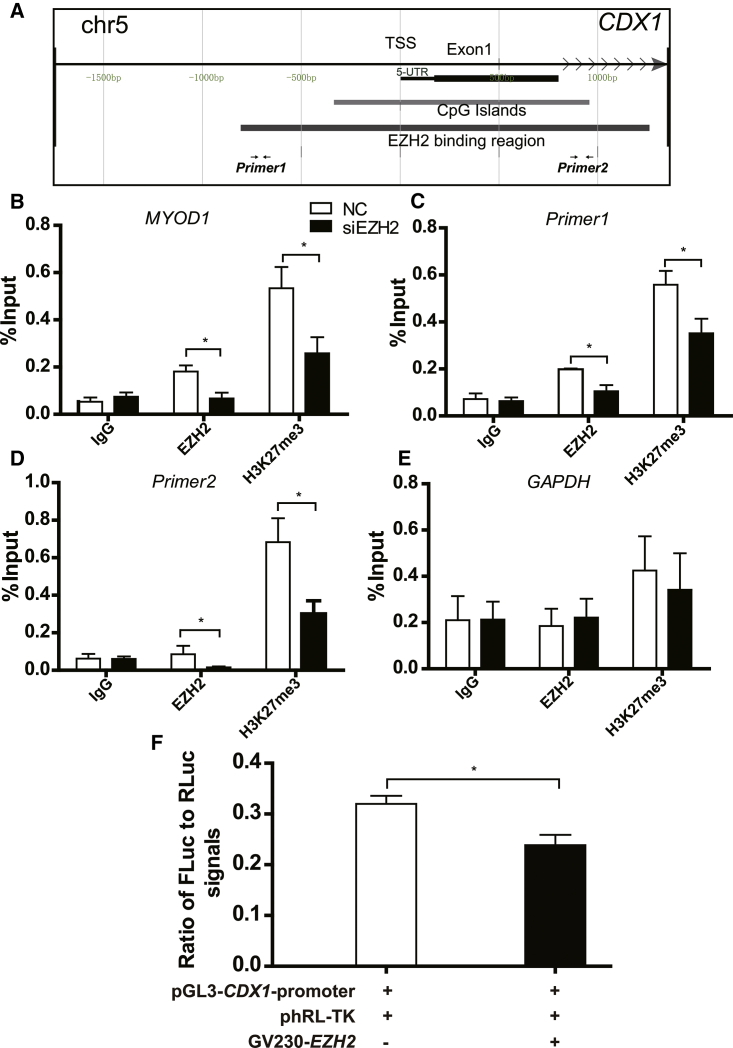
Figure 4.
EZH2 Regulated CDX1 Expression via Direct Binding to Its Promoter
(A) Schematic diagram of the CDX1 promoter region showing the location of CpG islands, the putative EZH2-binding region, and the spanning region of primers used for ChIP assay and PCR analysis. (B–E) Knockdown of EZH2 decreased the binding of both EZH2 and H3K27me3 on the promoter of CDX1. Immuno-precipitated DNA fragments were analyzed by qPCR with specific primer sets. Chromatin obtained from trophoblast cells was immune-precipitated using antibodies against EZH2, histone H3, and IgG. Each ChIP experiment was repeated three times. MYOD1 (B) was positive control; Primer1 (C) and Primer2 (D) were for EZH2; and GAPDH (E) was negative control. (F) Overexpression of EZH2 decreased the transcriptional activity of CDX1 promoter, as detected by dual-luciferase report assays. Approximately 2 kb upstream of the transcriptional start site (TSS) on the promoter of CDX1 was cloned into pGL3 vector. Three plasmids were cotransfected into trophoblast cells for 48 h. Fluorescence measurement was performed using dual-luciferase report assays. RLuc was used for the internal control. All data are reported as the mean ± SEM. *p < 0.05; **p < 0.01. FLuc, firefly luciferase; GAPDH, negative control; MYOD1, positive control; RLuc, Renilla luciferase.