Figure EV4. Characterisation of mito‐Rosella Caenorhabditis elegans and the effect of hV337M‐Tau on mitophagy in C. elegans neurons.
-
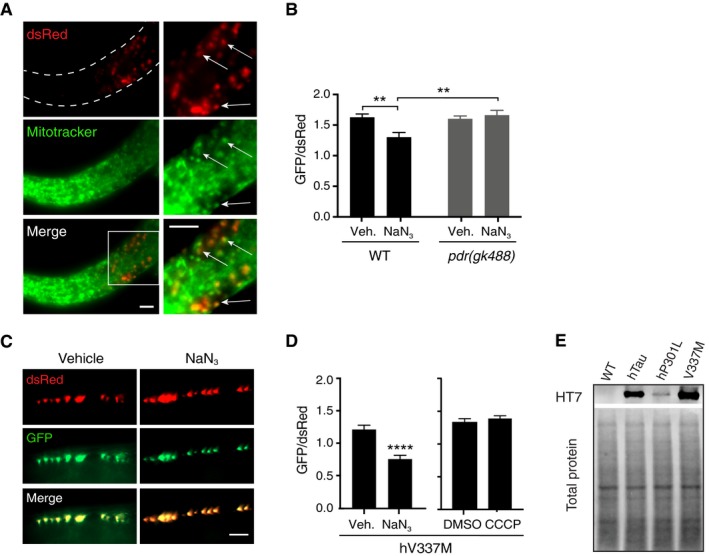
AMitotracker Deep Red staining illustrating colocalisation between Mitotracker (pseudo‐coloured green for better visualisation) and mito‐Rosella (dsRed). Scale bars = 5 μm.
-
BAnalysis of mitophagy in wild‐type worms and pdr‐1(gk488) mutant animals in response to NaN3. Data were analysed using two‐way ANOVA, revealing a main effect of genotype F(1, 106) = 6.415, P = 0.0128; a main effect of NaN3 treatment F(1, 106), P = 0.047; and a significant interaction effect F(1, 106), P = 0.0041; n = 26–29 animals/group.
-
C, DAnalysis of mitophagy in worms expressing hV337M‐Tau in response to NaN3 (Mann–Whitney U‐test, U = 25, P < 0.0001, n = 16, 19 for vehicle and NaN3, respectively), and CCCP treatment (unpaired t‐test, t = 0.8113, P = 0.4211, n = 25, 27 for DMSO and CCCP, respectively).
-
ETau expression in worm lysates detected with the human tau‐specific antibody HT7 (extended immunoblot from Fig 6I).
