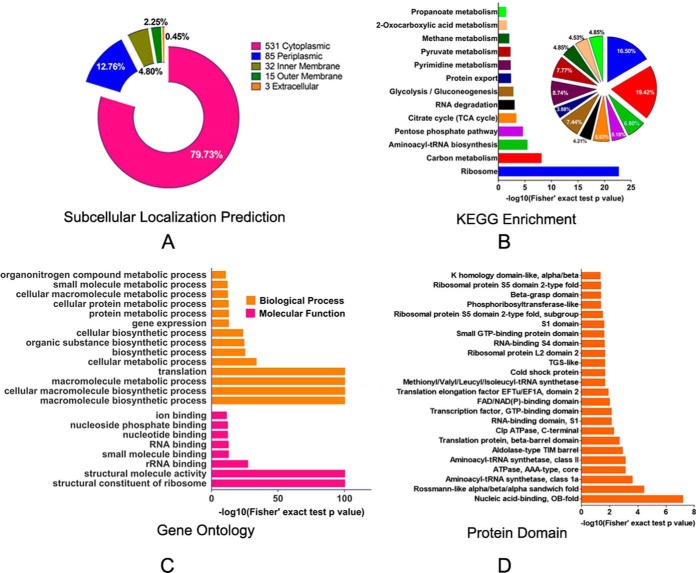
Fig. 2.
Subcellular prediction of proteins and functional classifications of succinylated-lysine proteins in A. hydrophila ATCC7966. A, Subcellular localization of the identified Ksucc proteins represented as a pie chart. B, KEGG pathway-based enrichment analysis of succinyl-proteins in A. hydrophila. The enrichment of identified proteins against all KEGG database proteins was used to identify enriched pathways via two-tailed Fisher's exact tests. Pathways with corrected p values < 0.05 were considered significantly enriched; C, GO-based enrichment analysis of identified proteins in A. hydrophila. A two-tailed Fisher's exact test was employed to test the enrichment of the identified proteins in each category against all of the database proteins. GOs with corrected p values < 0.05 were considered significantly enriched; D, Enrichment and analysis of domains related to succinylated proteins of A. hydrophila. For each category of proteins, the InterPro database was used to identify domains and a two-tailed Fisher's exact test was employed to assess the enrichment of identified proteins against all database proteins. Correction for multiple hypothesis testing was conducted using standard false discovery rate control methods. Domains with corrected p values < 0.05 were considered significantly enriched.