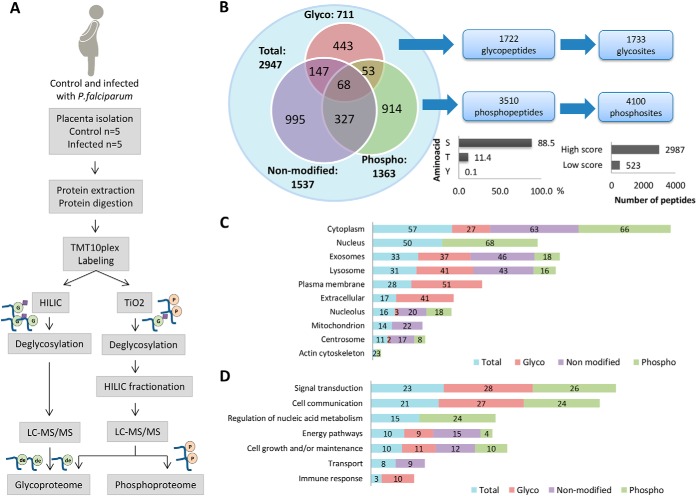
Fig. 3.
Integrated analyses of proteome, phosphoproteome and glycoproteome using isobaric labeling strategy in past P. falciparum infected and noninfected human placentas. A, Proteomics workflow for enrichment, identification, and quantification of phosphopeptides and glycopeptides in past P. falciparum infected and noninfected human placentas. B, Overall distribution of proteins identified in the proteome, phosphoproteome and glycoproteome fractions and total number of glycopeptides/glycosites and phosphopeptides/phosphosites identified by LC-MS/MS. The bar graphs represent % phosphosites in S, T or Y (left) and the number of phosphosites with high score (>0.99) and low score (<0.99). C, Percentages of proteins identified in the proteome, phosphoproteome and glycoproteome located in the indicated ten major organelles based on their gene ontology cellular components D, and seven major gene ontology biological processes.