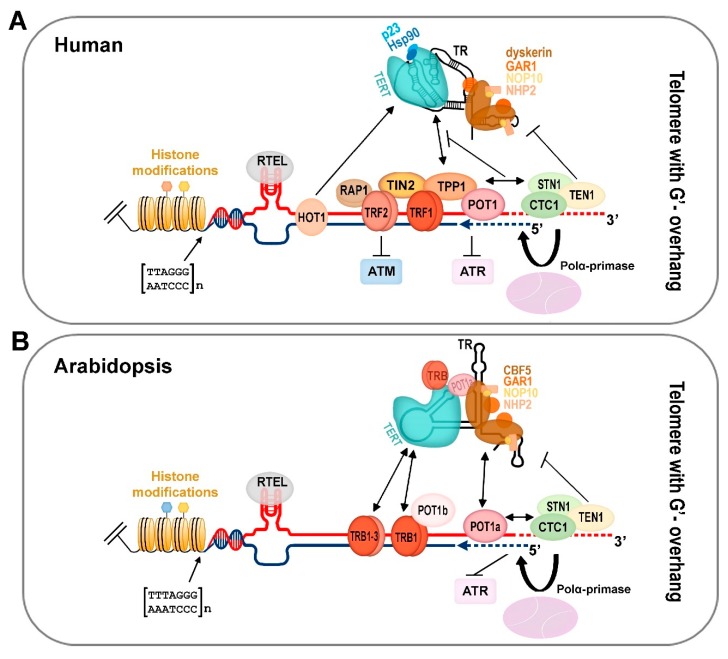
Figure 2.
An integrative schematic view of the human and plant terminal telomeric complex. (A) Human active telomerase is associated with Hsp90 and p23 chaperones as well as with TR associated conserved scaffold proteins of box H/ACA small nucleolar RNAs (dyskerin, NHP2, NOP10, GAR1). Mammalian shelterin proteins (TRF1/2, RAP1, TIN2, TPP1, and POT1) modulate access to the telomerase complex and the ATR/ATM-dependent DNA damage response pathway. The CST complex (CTC1-STN1-TEN1) affects telomerase and DNA polymerase α recruitment to the chromosomal termini, and, thus, coordinates G-overhang extension by telomerase with fill-in synthesis of the complementary C-strand (blue dashed line). G-quadruplexes, D-loops, and t-loops during telomere replication are resolved by RTEL helicase. HOT1 directly binds double strand telomere repeats and associates with the active telomerase. Telomere nucleosomes show a shorter periodicity than that in the other parts of chromosomes. For human telomere histone modifications, see Figure 3. (B) Arabidopsis telomerase is associated with TRB proteins as well as with POT1a that interacts with the dyskerin orthologue CBF5. Plants possess all orthologue proteins of conserved scaffold box H/ACA of small nucleolar RNAs (CBF5, GAR1, NOP10, NHP2). Moreover, TRB proteins interact with the telomeric sequence due to the same myb-like binding domain as that in mammalian TRF1/2. TRB proteins interact with TERT and POT1b, and, when localized at chromosomal ends, they are eligible to function as components of the plant shelterin complex. An evolutionarily conserved CST complex is suggested to coordinate the unique requirements for efficient replication of telomeric DNA in plants as well as in other organisms. In addition, plant RTEL contributes to telomere homeostasis. For the sake of clarity, only the situation in telomere with 3′ overhang is depicted. For plant telomere histone modifications, see Figure 3.