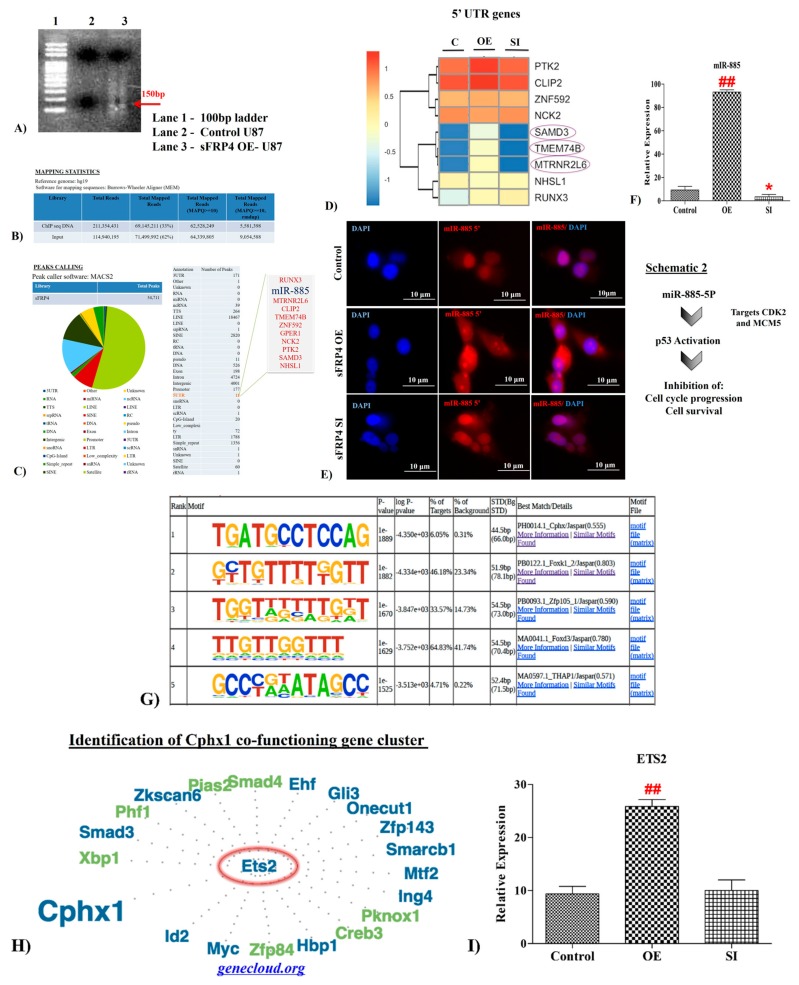
Figure 1.
(A–F). Apoptosis related genes identified in sFRP4 chromatin immunoprecipitation (ChIP) and pull-down sequencing analysis. Schematic 1 represents the sequence of steps of ChIP and downstream sequencing. ChIP DNA resolved on agarose gel indicated a 150 bp DNA band (A), ChIP mapping statistics by Burrow-Wheeler Aligner software indicated 5,581,398 mapped reads (B); peak calling analysis output using MACS2 software revealed 34,711 peaks related to secreted frizzled-related protein 4 (sFRP4) (C, left panel), categorization of peak identities represented by pie chart and table, analysis gene list present within 5′UTR (enlarged) indicated the presence of miRNA 885 (C, right panel), RNA sequencing data of 5′UTR revealed an upregulation of three 5′UTR genes in sFRP4 OE (OE) and downregulation in sFRP4 SI (SI) cells as indicated in the box (D), upregulation of active miR-885 in sFRP4 OE cells using an miR-885 5′LNA probe was detected as red fluorescence using a fluorescence microscope (scale bar = 10 μm) (E), quantitative RT-PCR indicated an over expression of miR885 in sFRP4 OE and downregulation in sFRP4 SI cells (F). Schematic 2 represents a model indicating the mode of action of miR-885 through its target genes CDK2 and MCM5 in cellular homeostasis via activation of p53. Results are mean ± SD of three independent experiments performed in triplicates (* p value < 0.05, # p value < 0.01, ## p value < 0.001). (G–I) HOMER de novo motif analysis and gene cluster analysis identifies senescence related genes in sFRP4 OE cells. Table representing LOGOS diagram of first five ranking genes motifs obtained by HOMER motif analysis software (G). Gene cluster analysis showing interlinking genes with Cphx1 gene (first ranked motif) using Gene cloud software identified the senescence associated gene, ETS2 (H). Relative mRNA expression analysis revealed ETS2 (Cphx1 co-functioning gene) was overexpressed in sFRP4 OE cells (I). Results are mean ± SD of three independent experiments performed in triplicates (*p value < 0.05, # p value < 0.01, ## p value < 0.001).