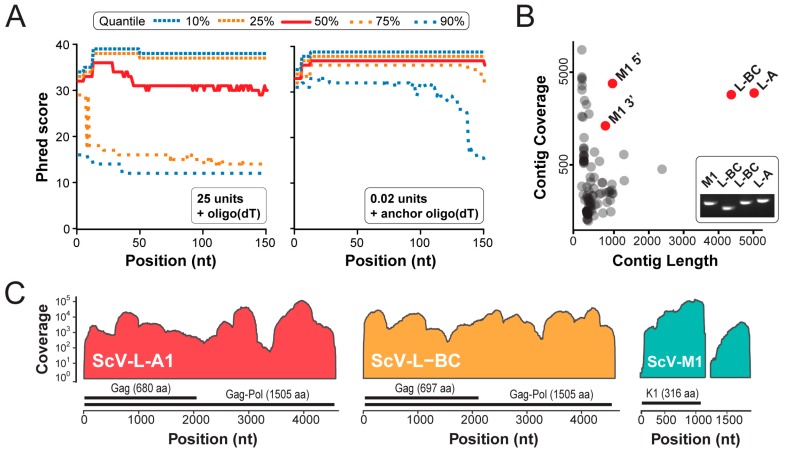
Figure 3.
Next-generation sequencing of totiviruses and dsRNA satellites from Saccharomyces cerevisiae strain BJH001. (A) Read quality (phred score) after Illumina QC shown along the length of the sequencing reads when using different concentrations of poly(A) polymerase and oligo(dT) or anchored oligo(dT) primers; 10%, 25%, 75%, and 90% quantile and median (50% quantile) read quality at each position along the reads are shown. (B) Sequence contigs after de novo assembled represented by contig coverage and contig length. BLAST analysis of the four contigs with the longest length and deepest coverage enabled their identification as totiviruses (ScV-L-A1 and ScV-L-BC) and a dsRNA satellite (ScV-M1), the latter was assembled as two separate contigs. Inset reverse transcriptase-PCR was used to confirm the presence of each type of dsRNA. Two primer pairs were used to amplify the ScV-L-BC. (C) Read depth coverage across the reference-assembled ScV-L-A, ScV-L-BC, and ScV-M1 contigs. Open reading frames present within each dsRNA are shown above the nucleotide position.