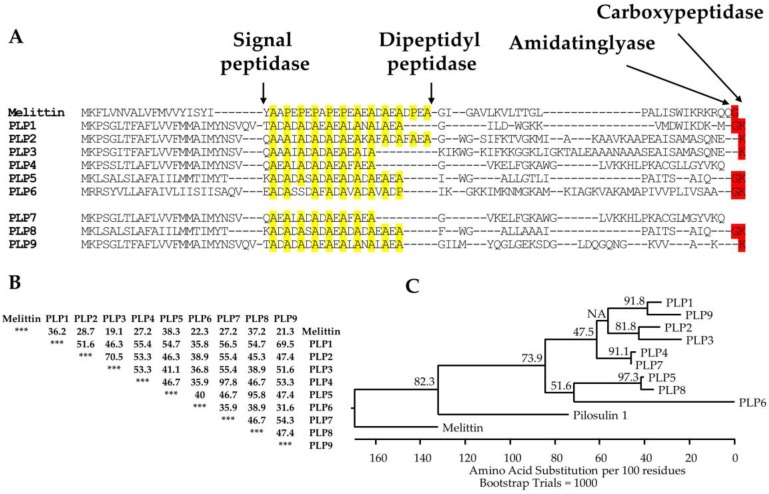
Figure 2.
Multiple alignment, identity matrix, and phylogenic analysis of pilosulin-like peptides. (A) The amino acid sequences of melittin and pilosulin-like peptides were aligned with ClustalW in Lasergene 12 (DNASTAR, Madison, WI, USA) and manually modified. Arrows indicate the putative processing and modification sites for signal peptidase, dipeptidyl peptidase, amidatinglyase, and carboxypeptidase. Proline and alanine residues in the spacer region between the signal and mature peptides of pilosulin-related peptides are highlighted in yellow. Nucleotide sequences for pilosulin-like peptides 7, 8, and 9 were assigned DDBJ/EMBL/GenBank Accession Numbers LC416796–LC416798, respectively. (B) Percentage amino acid sequence identities between melittin and pilosulin-like peptides are shown. (C) The alignment of pilosulin-like peptides, pilosulin 1, and melittin precursors by ClustalV in Lasergene 12 was used to construct a phylogenic tree using the neighbor-joining (NJ) method. The phylogenic tree rooted with the amino acid sequence of melittin. The numbers above the branches indicate the percentage of 1000 bootstrap replicates.