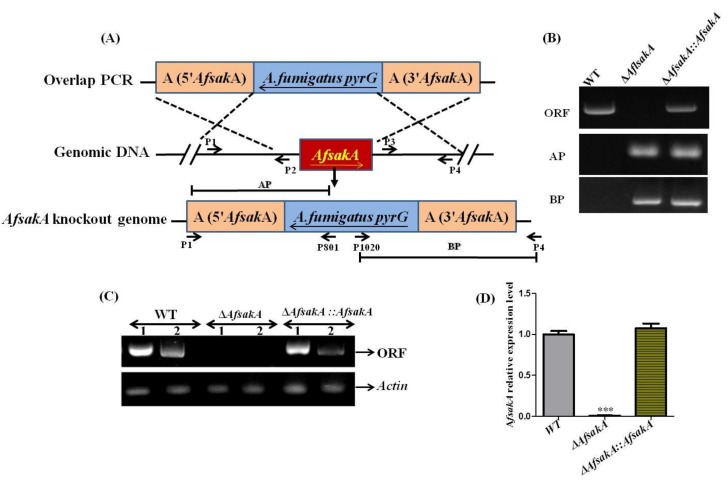
Figure 2.
Schematic illustration of AfsakA deletion and complementation strains of A. flavus. (A) Homologous recombination technique was applied for the deletion of AfsakA. The fragments 5′ UTR (untranslated region) (5′AfsakA), A. fumigatus pyrG and 3′ UTR (3′AfsakA) were each amplified with primer pairs of P1/P2, pyrG-F/pyrG-R and P3/P4 and fused together with primer pairs P7/P8. The fusion PCR product was transferred into CA14 (PyrG, Δku70) in order to produce ΔAfsakA (Δku70; pyrG; ΔAfsakA::pyrG). (B) The results of the PCR analysis of ΔAfsakA and ∆AfsakA::AfsakA. ORF (open reading frame), AP and BP fragments were amplified with a couple of primers P9/P10, P1/P801 and P4/P1020 respectively. (C) Verification of AfsakA deletion and complementary strains with reverse transcription-PCR. Actin gene was used as an endogenous control. Lane 1 used gDNA and lane 2 used cDNA as template. (D) qRT-PCR analysis of the expression level of AfsakA gene in ∆AfsakA mutant, WT (wild-type) and ∆AfsakA::AfsakA strains. The line bar in every column in (D) indicates standard errors of four replicates and the asterisks show significant difference between the knockout mutant and other strains (WT and complementation strain) (*** p ≤ 0.001).