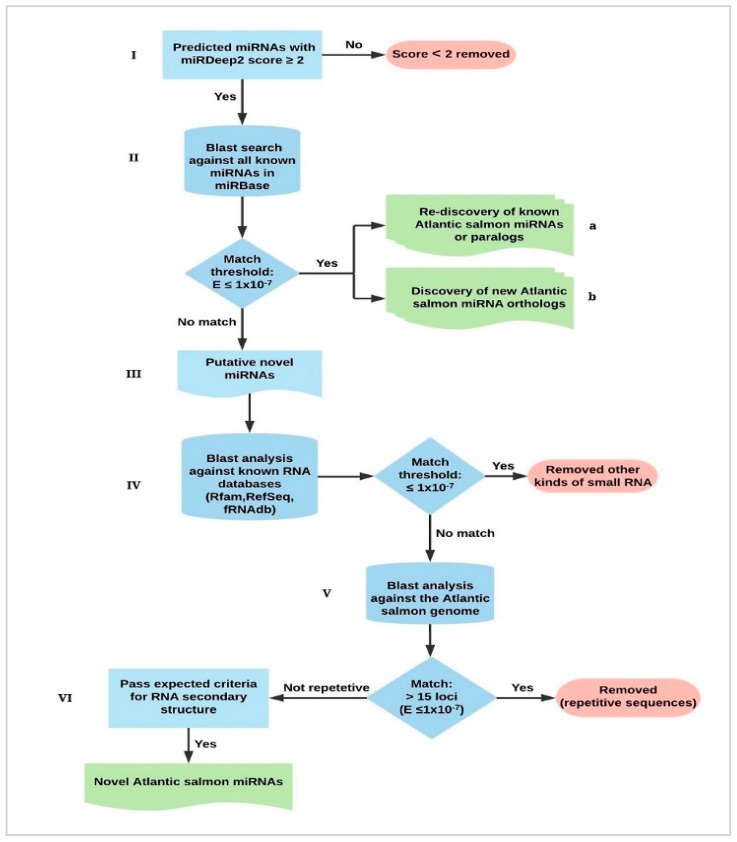
Figure 1.
Identification and characterization of Atlantic salmon microRNAs (miRNAs). (I) Candidate miRNAs with miRdeep2 score ≥ 2 were regarded as putative miRNAs. (II) miRNA precursors were used as input for BLAST search against miRBase to identify; (a) Atlantic salmon miRNAs already in the database and new paralogs, (b) Orthologues of other teleost miRNAs not discovered in prior studies of Atlantic salmon. (III) Candidate miRNAs with no significant matches (e ≤ 1 × 10−7) to miRBase were considered as putative novel Atlantic salmon miRNAs. (IV) Putative novel miRNA precursor sequences were BLAST analyzed against other RNA databases (Rfam, NCBI RefSeq and fRNAdb) to remove those that were other kinds of small RNA. (V) Sequences with no matches in these databases were further used for BLAST analysis against the Atlantic salmon reference genome sequence. Significant matches (e ≤ 1 × 10−7) to more than 15 loci in the Atlantic salmon genome were annotated as interspersed repeats and removed. (VI) The remaining putative novel miRNAs were compared against expected features of precursors and their mature miRNAs. The candidates that passed all these criteria were regarded as novel Atlantic salmon miRNAs.