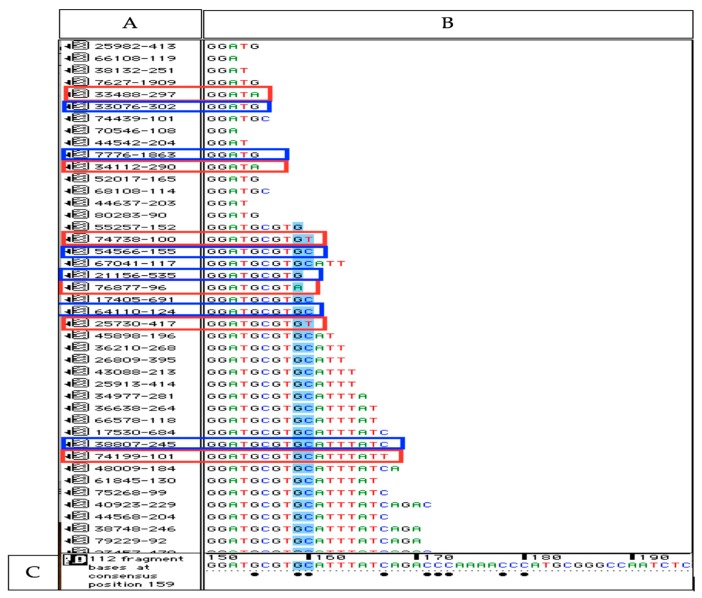
Figure 2.
Illustration of aligned reads to the reference (18S rRNA) applying Sequencher software. (A) This window shows each collapsed read identified by a unique number followed by the count numbers of this particular read. (B) The sequence of each collapsed read is given in this window (C) The sequence at the bottom is the reference sequence (18S rRNA). Bullets below bases of the reference indicate that some of the reads differ from the reference at these basepair positions (erroneous sequence variants, ESVs). All collapsed reads shown in red boxes have 3’ ESVs. The collapsed reads shown in blue boxes are reads with correct bp in their 3’ end position. The ratio of reads with 3’ ESVs compared to reads with correct 3’ end bp’s in this short part of the sequence (bp 150–bp 170) was 0.10. The other reads below 3’ ESV were all identical to reference at these base positions (e.g., bases in blue color) verifying that the misaligned bases are 3’ ESVs not polymorphic variants.