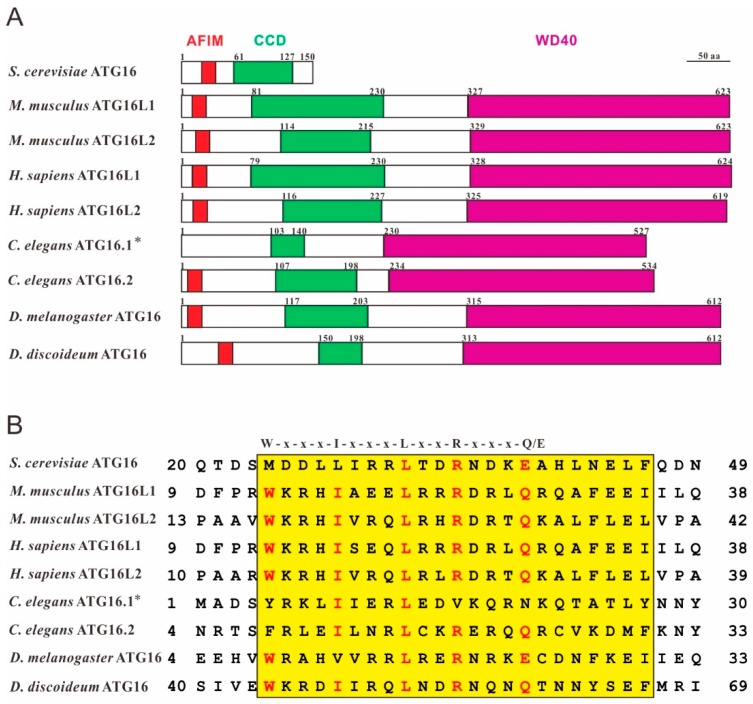
Figure 1.
ATG16 shares high structural similarity in different species. (A) Domain structures of Saccharomyces cerevisiae ATG16 (NP_013882.1), Homo sapiens ATG16L1 (NP_001350671.1) and ATG16L2 (NP_203746.1), Mus musculus ATG16L1 (NP_001192320.1) and ATG16L2 (NP_001104581.1), Caenorhabditis elegans ATG16.1 (NP_508768.1) and ATG16.2 (NP_495299.2), Drosophila melanogaster ATG16 (NP_001138124.2) and Dictyostelium discoideum ATG16 (XP_643673.1). Conserved domains were predicted using SMART (http://smart.embl-heidelberg.de/). AFIM (ATG5-interacting motif), red; CCD (coiled-coil domain), green; WD40 (tryptophan-aspartic acid, WD40, repeat domain), violet. * the sequence used here lacks the N-terminal 51 amino acids because it was shown that the C. elegans atg-16.1 gene encodes 527 amino acids and not, as originally predicted 578 amino acids [18]. (B) Sequence alignment of the AFIM regions of different ATG16 proteins using the Multiple Sequence Alignment program at the NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The highly conserved AFIM residues are denoted on the top [19]. Red letters highlight conserved key residues of the AFIM and yellow shading indicates the highly conserved AFIM region between species.