Figure 2.
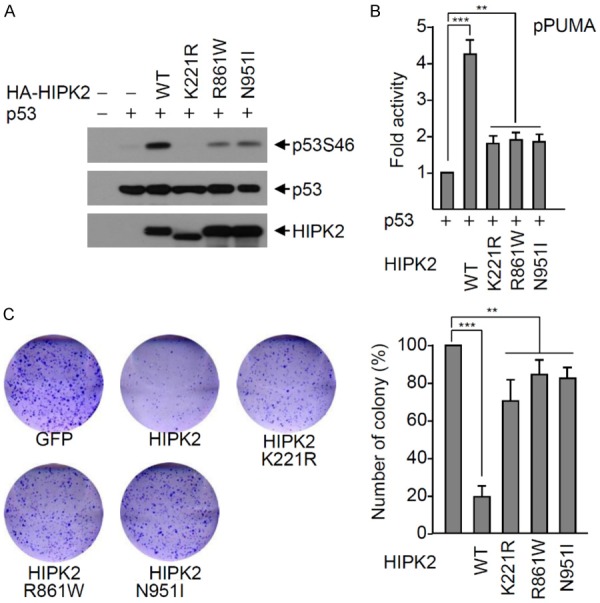
The HIPK2 R861W and N951I mutants do not phosphorylate p53 at Ser46. A. H1299 cells were transfected with p53 and either wild-type HA-HIPK2 or a HIPK2 mutant (catalytically inactive K221R mutant, R861W or N951I) expression plasmid. p53 Ser46 phosphorylation was determined with Western blotting using a p53Ser46 phospho-specific antibody. Expression levels of p53 and HIPK2 were determined by Western blotting using anti-p53 and anti-HA antibody, respectively. B. The p53 expression plasmid and each reporter plasmid (PUMA gene promoter) was transfected into H1299 cells with either wild-type HIPK2 or an HIPK2 mutant (K221R, R861W or N951I mutant). Twenty-four hours after transfection, luciferase activities were measured using the Luciferase Assay System (Promega). Transfection efficiency was normalized against β-galactosidase expression. Experiments were repeated at least three times. The data were statistically analyzed by one-way ANOVA followed by Bonferroni’s multiple comparison test (***P < 0.001, **P < 0.01). Bars represent the mean ± SD of three independent experiments. C. The HIPK2 R861W and N951I mutants are defective in p53-mediated induction of apoptosis. Wild-type GFP-HIPK2 or a GFP-HIPK2 mutant was transfected into RKO cells, and transfected cells were selected with G418 for two weeks. Colonies were visualized by staining with crystal violet and counted. The number of colonies was determined as the mean ± SD of three independent experiments, and presented as the relative percentage compared to the colony numbers of the negative control. The data were statistically analyzed by one-way ANOVA followed by Bonferroni’s multiple comparison test (***P < 0.001, **P < 0.01). A representative example is shown.
