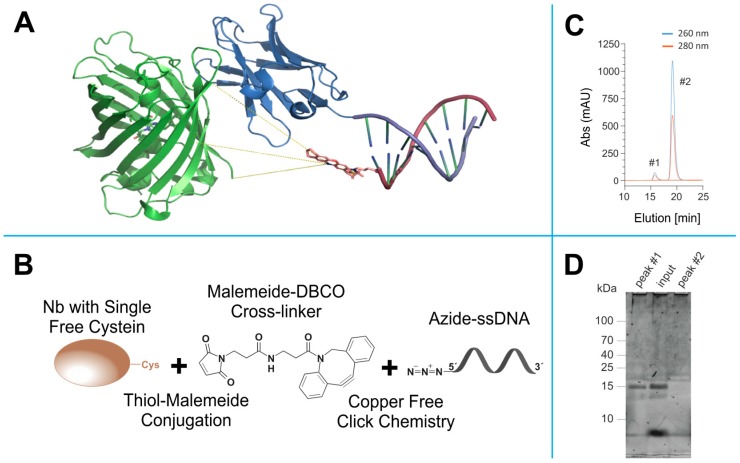
Figure 2.
Click- and thiol-based strategy to conjugate nanobodies to a docking DNA strand for DNA-PAINT. (A) Anti-GFP nanobody (blue) bound to EGFP (green). The nanobody is modified with a docking stand with a complementary Atto655-labelled Imager strand attached (EGFP: Nanobody complex extracted from (PDB: 3K1K), DNA strand, and Atto655 were generated using ChemDraw (CambridgeSoft, Cambridge, MA, USA) and ensembled using PyMOL Molecular Graphics System (Schrödinger, LLC, Ney York, NY, USA). The yellow lines represent three estimated distances (theoretical estimates: 3.1 nm, 3.3 nm and 3.4 nm) of the fluorophore to the protein of interest (POI) extracted from the in silico model. (B) Scheme representing the orthogonal coupling strategy of docking DNA strand to the nanobody. (C) Example of the size-exclusion chromatography (SEC) for the separation of DNA-coupled nanobody (#1) from the excess of azide-functionalized docking strand (#2). (D) Example of the SDS-PAGE of fraction collected from the SEC run, post stained with SYBR gold, which reports DNA on the gel. Peak #1 collected from SEC shows a prominent band matching the expected molecular weight of nanobody coupled to the docking strand (~15 kDa). Peak #2 lacks the band at the nanobody molecular weight, suggesting that the SEC peak contains only the un-reacted excess of docking oligonucleotide.