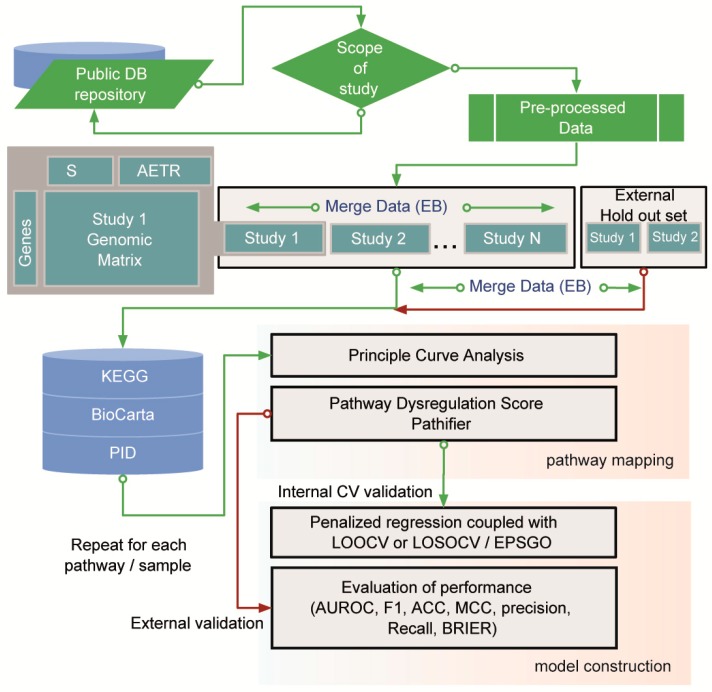
Figure 1.
Pipeline for performing a meta-analysis-derived, multivariate model for personalized pathways in acquired epidermal growth factor inhibitor tyrosine kinase inhibitor (EGFR TKI) resistance (AETR). The pipeline consists of three main parts: cross study normalization, pathway mapping, and prediction model construction. The study cohort was preprocessed and categorized into an internal training/validation study set (N) and an external validation study set (M). For cross-study normalization, an empirical Bayes (EB) method was used. Pathway mapping for each individual sample was conducted using a Pathifier algorithm and public pathway databases (KEGG, BioCarta, and PID). The regularized regression model was built using elastic net. The optimal values of the hyper-parameters α and λ for elastic net regression were obtained from robust cross validation (leave-one-study-out cross validation (LOSOCV) or leave-one-out cross validation (LOOCV)) with Efficient Parameter Selection via Global Optimization (EPSGO) algorithm. S, sensitive.