Abstract
Accumulating evidence indicates that long non-coding RNAs (lncRNAs) play a key role in the development of many human cancers. MT1JP is a lncRNA that is reportedly involved in gastric cancer development, but a biological role and mechanism for MT1JP in breast cancer is unknown. Here, we found that MT1JP expression was significantly down-regulated in breast cancer tissues and cell lines, and that decreased MT1JP expression was associated with breast cancer progression and poor survival of breast cancer patients. Additionally, we found that overexpression of MT1JP in breast cancer cells significantly inhibited cell proliferation and invasion, and also enhanced the cisplatin sensitivity of breast cancer cells. We then investigated a possible mechanism for these results, finding that MT1JP binds to and negatively regulates miR-24-3p, which is known to be an oncogene in some human cancers. Our rescue experiments showed that the tumor suppressive and cisplatin-sensitizing functions of MT1JP were mediated by negative regulation of miR-24-3p. Finally, western blot results showed that MT1JP inhibited the Wnt/β-catenin signaling pathway. Collectively, our data indicate that MT1JP functions as an anti-tumor lncRNA, enhances cisplatin sensitivity in breast cancer, and may serve as a novel diagnostic and therapeutic marker of breast cancer.
Keywords: MT1JP, breast cancer, proliferation, cisplatin, miR-24-3p
Introduction
Breast cancer is one of the most common malignancies that affects women worldwide, and its incidence is gradually increasing. It is also the second most common cause of cancer-related mortality in women [1,2]. Despite great advances in basic research and clinical treatment, breast cancer still has a poor prognosis [3,4]. The unfavorable prognosis can be attributed to incomplete diagnoses, high recurrence, and treatment resistance, as well as the adverse effects of comprehensive therapies [5-7]. Therefore, it is important to understand the potential molecular mechanisms involved in breast cancer development and identify novel diagnosis and therapeutic biomarkers of breast cancer.
Long non-coding RNAs (lncRNAs) are defined as a class of non-coding RNA longer than 200 nucleotides. Although the absence of protein coding previously led them to be considered transcriptional “noise” without any biological function [8], increasing evidence has shown that lncRNAs have important roles; these roles include acting as powerful transcriptional and post-transcriptional mediators in processes such as the initiation and progression of breast cancer [9-11]. The lncRNA MT1JP is located on chromosome 16 in a cluster that consists of several homologous protein-coding genes in the metallothionein family. MJ1JP was first reported as a tumor suppressor that regulated the p53 protein expression level, thereby regulating the p53-related signaling pathway [12]. Later, MT1JP was found to be down-regulated in gastric cancer tissues and cell lines. Low MT1JP expression strongly correlated with lymphatic metastasis, advanced Tumor, Node, Metastasis (TNM) stage, and shorter survival times for gastric cancer patients. Furthermore, MT1JP overexpression inhibits gastric cancer cell proliferation, migration, and invasion; promotes gastric cancer cell apoptosis in vitro; and suppresses tumor growth and metastasis in vivo [13,14]. However, the expression, function, and potential molecular mechanism of MT1JP in breast cancer remains unclear.
In this study, we measured the expression of MT1JP in breast cancer tissues and corresponding adjacent normal tissues, and explored its function in breast cancer cell lines. We also investigated the roles of MT1JP in cisplatin (DDP) sensitivity. In our investigation of the mechanism by which MT1JP regulated breast cancer cell line proliferation, invasion, and cisplatin sensitivity, we found that MT1JP mediated these effects by negatively regulating miR-24-3p. To the best of our knowledge, these data are the first indication that MT1JP plays an important tumor-suppression role in breast cancer.
Materials and methods
Human tissue samples
Between January 2006 and December 2011, 56 paired samples of breast cancer tissue and adjacent normal breast tissues were collected from patients undergoing resection surgery at the Department of Oncology, Shanghai East Hospital. None of the patients received any preoperative anti-cancer treatment, such as chemotherapy or radiation before sample collection. All samples from resection surgery were frozen and stored in liquid nitrogen until use. We performed this study with the approval of the Ethic and Research Committees of Shanghai East Hospital and in accordance with the Declaration of Helsinki Principles. Written informed consents were obtained from all subjects. The clinical characteristics of all the patients are summarized in Table 1.
Table 1.
Expression of MT1JP in relation to pathologic and clinical features of breast cancer patients
| Clinico-pathological features | N2 | MT1JP expression | P value | |
|---|---|---|---|---|
|
| ||||
| H | L | |||
| All | 56 | 31 | 25 | |
| Age (years) | 0.863 | |||
| < 60 | 32 | 18 | 14 | |
| ≥ 60 | 24 | 13 | 11 | |
| Tumor size | 0.027 | |||
| < 2 cm | 23 | 15 | 8 | |
| ≥ 2 cm | 33 | 16 | 17 | |
| ER status | 0.417 | |||
| Negative | 30 | 17 | 11 | |
| Positive | 26 | 14 | 12 | |
| PR status | 0.251 | |||
| Negative | 27 | 16 | 11 | |
| Positive | 29 | 15 | 14 | |
| Her-2 status | 0.330 | |||
| Negative | 44 | 24 | 20 | |
| Positive | 12 | 7 | 5 | |
| Lymph node metastasis | 0.493 | |||
| Negative | 28 | 16 | 12 | |
| Positive | 28 | 15 | 13 | |
| Ki-67 | 0.067 | |||
| Negative | 25 | 15 | 10 | |
| Positive | 31 | 16 | 15 | |
| TNM stage | 0.032 | |||
| I-II | 49 | 31 | 18 | |
| III | 7 | 0 | 7 | |
Cell culture
The breast epithelial cell line MCF-10A and human breast cancer cell lines MDA-MB-231, SKBR-3, MCF-7, and MDA-MB-468 were purchased from American Type Culture Collection (ATCC, USA). They were cultured in Dulbecco’s modified Eagle’s medium (DMEM), and supplemented with 10% fetal bovine serum, 50 U/ml penicillin, and 0.1 mg/ml streptomycin. All cells were maintained at 37°C in a 5% CO2 humidified atmosphere.
Constructs, synthesized oligos, and transfection
The MT1JP overexpression plasmid was purchased from GenePharm (Shanghai, China). The antimiR-24, miR-24 mimic, and scrambled siRNA (si-NC) were also purchased from Shanghai Gene Pharma Co. Ltd (Shanghai, China). The sequence was amplified and inserted into the pcDNA3.1 (+) vector at the BamH1 sites. For transfection, the MDA-MB-231 and SKBR-3 cells were grown in 6-well plates to 50-70% confluence, then were transfected with the indicated molecules using Lipofectamine 3000 (Thermo) according to the manufacturer’s instructions. Forty-eight hours after transfection, cells were ready for the following experiments.
RNA extraction and real-time polymerase chain reaction (PCR)
Total RNA from tissues and cells was isolated using Trizol reagent (Invitrogen, Carlsbad, USA) according to the manufacturer’s instructions. RNA was reverse transcribed with the PrimeScript RT Reagent Kit (Invitrogen, USA), quantitative reverse transcription PCR (qRT-PCR) was performed using SYBR Premix Ex Taq (TaKaRa, China), following the manufacturer’s instructions. GAPDH was used as an internal lncRNA control. The primer sequences were as follows: MT1JP, forward: 5’-TACCGAGCTCGGATCCTTGCGGTCTCTCCATTTATCG-3’, reverse: 5’-TACCGAGCTCGGATCCTTGCGGTCTCTCCATTTATCG-3’; GAPDH, forward: 5’-GCACCGTCAAGGCTGAGAAC-3’, reverse: 5’-TGGTGAAGACGCCAGTGGA-3’. We performed qRT-PCR using the ABI PRISM 7500 PCR System (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions.
Cell proliferation assay
After transfection with the empty vector or pcDNA-MT1JP, we used the Cell Counting Kit-8 (CCK-8; Dojindo Molecular technologies, Inc., Kyushu, Japan) according to the manufacturer’s protocol to measure cell proliferation. Briefly, we plated 2000 cells into 96-well plates. We added 10 µl CCK-8 to the wells and measured the quantity of formazan formed at 450 nm absorbance following the instructions provided.
Colony formation assay
For the colony formation assay, we seeded transfected cells in 6-well plates at 500 cells per well and maintained these in media with 10% FBS. Two weeks later, we fixed cells with 4% paraformaldehyde and stained with crystal violet. Colonies with diameters of more than 1.5 mm was counted.
Cell invasion assay
The invasion assays were performed in Transwell chambers (Costar, Massachusetts, USA) with matrigel coated membranes, according to the manufacturer’s instruction. We added 1 × 105 cells suspended in 300 µl serum-free medium to the upper compartment of inserts in the 24-well plate, and added 800 µl DMEM supplemented with 10% FBS to the lower compartment. After 24 h incubation, non- penetrating cells on the upper surface of the membrane were removed with a cotton swab, then the membrane in the lower chamber was fixed with 4% formaldehyde and stained with 1% crystal violet. We quantified the number of cells that invaded through the membrane under a microscope for at least five fields. Experiments were performed in triplicate.
Dual-luciferase reporter assay
First, we co-transfected a reporter plasmid containing MT1JP wild type (wt) or MT1JP mutant (mut) with the miR-24 mimic or the miR-NC into MDA-MB-231 cells. Then, the reporter plasmid and the internal control plasmid containing renilla luciferase were transfected using Lipofectamine 3000 (Invitrogen). After 48 h, we examined the luciferase activity using the dual-luciferase reporter gene assay system (Promega, Madison, USA). Renilla luciferase activity was normalized to firefly luciferase activity.
Western blot
We performed western blots as previously described [15]. After incubation with the secondary antibody, we visualized protein bands using the enhanced chemiluminescence detection kit (Thermo Fisher Scientific, Waltham, MA, USA). The primary antibodies used in this study were: β-catenin, cyclin D1, c-myc, and β-actin. All antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz).
Statistical analysis
All results are shown as mean ± SD and were analyzed using GraphPad Prism 5 (GraphPad Software, USA) from at least three independent experiments. The chi-square tests explored the associations between MT1JP level and clinico-pathological factors. We used the Kaplan-Meier method to calculate the survival curve and the log-rank test to determine statistical significance. The differences between groups were analyzed using Student’s t-test. Data were considered to be statistically significant when P < 0.05.
Results
MT1JP expression was down-regulated in breast cancer tissues and cell lines
To investigate the role of MT1JP in breast cancer, we first used qRT-PCR to examine the relative expression of MT1JP in 56 paired samples of breast cancer tissues and adjacent non-tumor tissues. As shown in Figure 1A, the expression level of MT1JP was markedly down-regulated in breast cancer tissues compared to expression in adjacent non-tumor tissues. We than measured the MT1JP expression in four human breast cancer cell lines (MDA-MB-231, SKBR-3, MCF-7, and MDA-MB-468) and compared it to expression in a normal breast epithelial cell line, MCF-10A. Results showed that breast cancer cell lines had lower MT1JP expression than a normal breast cell line (Figure 1B). These data indicate that MT1JP expression is down-regulated both in human breast tissues and breast cancer cell lines.
Figure 1.
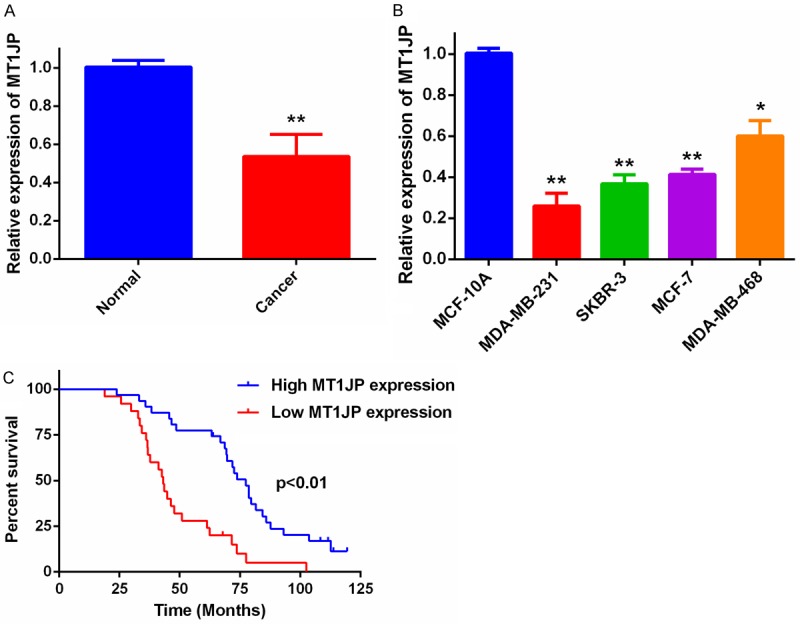
Relative MT1JP expression levels in breast cancer and the clinical significance. A. Relative MT1JP expression in 56 paired samples of breast cancer tissue and adjacent non-tumor tissues by qRT-PCR analysis. **P < 0.01. B. MT1JP levels in breast cancer cells compared with levels in normal MCF-10A breast epithelial cells. *P < 0.05, **P < 0.01. C. Kaplan-Meier survival curves for patients with breast cancer tissues expressing low and high levels of MT1JP.
Then, we further investigated the correlation between MT1JP expression and the clinico-pathological features of 56 breast cancer patients (Table 1). Our results show that down-regulation of MT1JP expression is significantly associated with tumor size (P = 0.027) and TNM stage (P = 0.032), but there is not significantly associated with age, estrogen receptor (ER) status, progesterone receptor (PR) status, Her-2 status, or Ki-67. In addition, Kaplan-Meier survival curves (Figure 1C) showed that breast cancer patients with higher MT1JP expression have significantly better overall survival than the lower expression group.
MT1JP overexpression impairs proliferation and colony formation of MDA-MB-231 and SKBR-3 cells
Because MT1JP expression was lower in MDA-MB-231 and SKBR-3 cells, we generated a MT1JP-overexpressing plasmid and performed overexpression experiments in these two cell lines. The results showed that the expression was upregulated, compared to cells with an empty vector (Figure 2A). CCK-8 assays showed that the MT1JP overexpression significantly impaired the proliferative abilities of MDA-MB-231 and SKBR-3 cells (Figure 2B and 2C). Similarly, MT1JP overexpression suppressed the colony formation abilities of MDA-MB-231 and SKBR-3 cells (Figure 2D and 2E).
Figure 2.

MT1JP overexpression inhibits proliferation of breast cancer cells. A. The Expression levels of MT1JP in MDA-MB-231 and SKBR-3 cells after transfection with pcDNA-MT1JP or empty vector. **P < 0.01. B. The CCK-8 assay showing effects of MT1JP overexpression on MDA-MB-231 cell proliferation. **P < 0.01. C. The CCK-8 assay showing effects of MT1JP overexpression on SKBR-3 cell proliferation. *P < 0.05, **P < 0.01. D. Effect of MT1JP overexpression on MDA-MB-231 colony formation and cell proliferation. *P < 0.05. E. Effect of MT1JP on SKBR-3 colony formation and cell proliferation. **P < 0.01.
MT1JP overexpression inhibits breast cancer cell invasion
To further examine the role of MT1JP in breast cancer cell invasion, we performed Transwell assays using MDA-MB-231 and SKBR-3 cells. We found that overexpression of MT1JP impaired the invasion abilities of MDA-MB-231 and SKBR-3 cells relative to those of the empty vector group (Figure 3).
Figure 3.
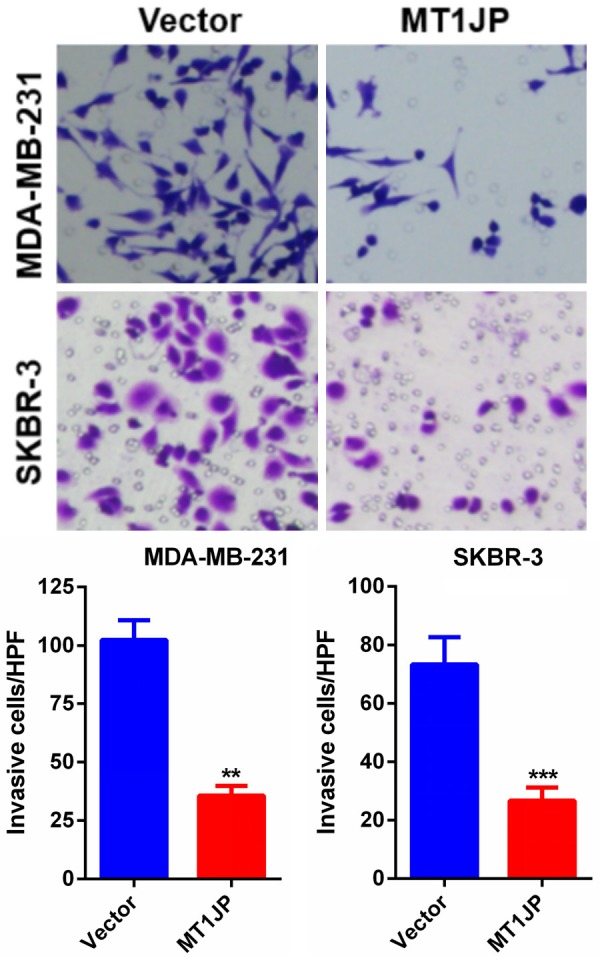
MT1JP overexpression suppresses MDA-MB-231 and SKBR-3 cells invasion. The Transwell assay showing effects of MT1JP overexpression on MDA-MB-231 and SKBR-3 cells invasion. **P < 0.01, ***P < 0.001.
MT1JP enhances cisplatin sensitivity
To determine the effect of MT1JP on the cisplatin sensitivity of breast cancer cells, we established two DDP-resistant breast cancer cells (MDA-MB-231/DDP and SKBR-3/DDP). We found higher IC50 values in MDA-MB-231/DDP and SKBR-3/DDP cells than in corresponding parental cell lines MDA-MB-231 and SKBR-3, indicating an increased resistance to DDP (Figure 4A). We performed qRT-PCR to measure MT1JP expression in parental and DDP-resistant breast cancer cells. We observed significantly reduced MT1JP expression in MDA-MB-231/DDP and SKBR-3/DDP cells (Figure 4B). Then, we performed overexpression experiments for MT1JP in MDA-MB-231/DDP and SKBR-3/DDP cells, and found that expression of MT1JP was upregulated after transfection with pcDNA-MT1JP 3 (Figure 4C). The CCK-8 results showed that MT1JP overexpression in MDA-MB-231/DDP and SKBR-3/DDP cells resulted in significantly lower IC50 values than those of the control group (Figure 4D). Together, these data suggest that MT1JP expression induces DDP sensitivity in DDP-resistant breast cancer cells.
Figure 4.

MT1JP overexpression enhances the cisplatin sensitivity of cisplatin-resistant CRC cells. A. The differential expressions of MT1JP in MDA-MB-231 and SKBR-3 cells and the cisplatin-resistant MDA-MB-231/DDP and SKBR-3/DDP cells. B. The expression level of MT1JP in MDA-MB-231/DDP and SKBR-3/DDP cells after transfection with pcDNA-MT1JP or empty vector. C. Effect of MT1JP on cisplatin sensitivity of MDA-MB-231/DDP cells. D. Effect of MT1JP on cisplatin sensitivity of SKBR-3/DDP cells.
Identification of miR-24-3p as a target of MT1JP
Because it is reported that MT1JP functions as a competing endogenous RNA (ceRNA) for specific miRNAs, we used RNAhybird2 and FINDTR3 to search for miRNAs that complementary base pair with MT1JP. We found five miRNAs that MT1JP could potentially target, then we compared the expression levels of these five miRNAs in MT1JP overexpression cells and in control cells. We found that miR-24-3p expression was most affected by MT1JP overexpression (Figure 5A). The dual-luciferase reporter assay confirmed the relationship between MT1JP and miR-24-3p. Results showed that cells co-transfected with pLUC-MT1JP-wild type and the miR-24-3p mimic (the MT1JP-wt + miR-24-3p group) had significantly lower luciferase activity than cells co-transfected with pLUC-MT1JP-wild type and the miR-NC (the MT1JP-wt + NC group). In contrast, we found no significant difference in the relative luciferase activity between the MT1JP-mut + miR-24-3p and the MT1JP-mut + miR-NC groups (Figure 5B and 5C). In addition, we examined miR-24-3p expression in 56 paired samples of breast cancer tissue and adjacent normal tissue by qRT-PCR and found that miR-24-3p expression was significantly higher in breast cancer tissues than in adjacent normal tissues (Figure 5D). Furthermore, the expression of miR-24-3p was negatively associated with MT1JP expression in breast cancer tissues (Figure 5E). These data strongly suggest that MT1JP targets and negatively regulates miR-24-3p expression in breast cancer tissues.
Figure 5.
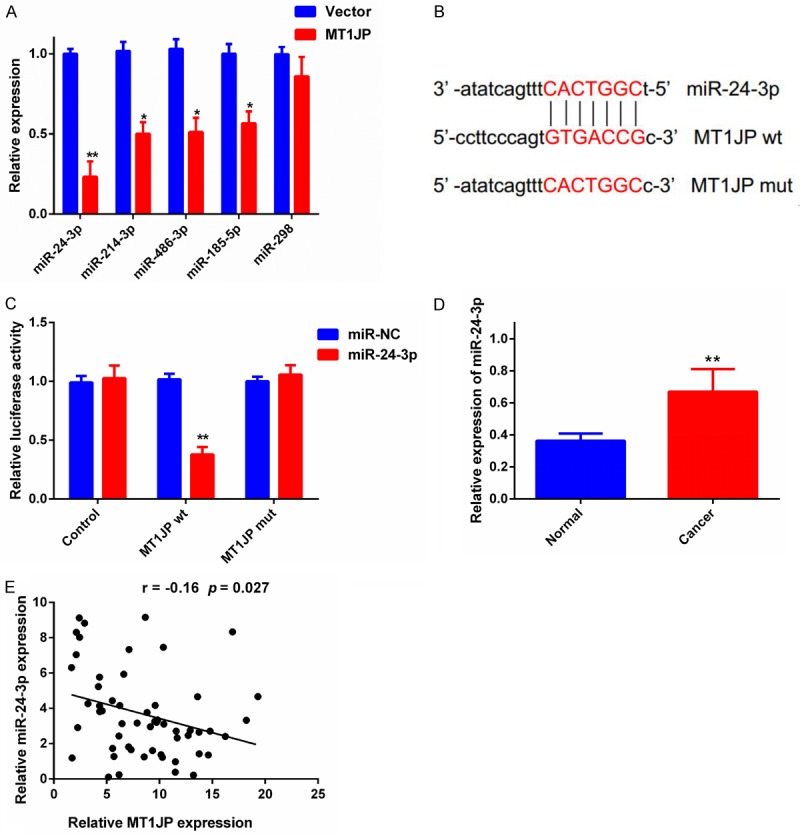
MiR-24-3p is a target of MT1JP. A. qRT-PCR to measure the expression of nine miRNAs in MT1JP overexpression and in control cells. *P < 0.05, ** P < 0.01. B. Putative complementary sites between MT1JP and miR-24-3p. Mutations were generated in the MT1JP nucleotides complementary to miR-320a. C. Luciferase activity in MDA-MB-231 cells co-transfected with the miR-24-3p mimic or miR-NC and in control luciferase reporters without the MT1JP wt or MT1JP mut insert. ** P < 0.01. D. qRT-PCR was used to measure miR-24-3p expression in 56 pairs of breast cancer tissue and adjacent normal tissue. ** P < 0.01. E. The correlation between MT1JP and miR-24-3p was evaluated.
The tumor suppressive and cisplatin-sensitized function of MT1JP in breast cancer cells is dependent on miR-24-3p
Finally, we performed rescue experiments to determine whether MT1JP influenced breast cancer cell proliferation, invasion, and cisplatin sensitivity in a miR-24-3p dependent manner. Our results in Figure 6A confirmed that miR-NC or miR-24-3p mimic was stably transfected into MDA-MB-231 cells that were already transfected with pcDNA-MT1JP. CCK-8 assays showed that the MT1JP overexpression-induced suppression of proliferation in MDA-MB-231 cells was partially abolished in the presence of the miR-24-3p mimic (Figure 6B). The presence of the miR-24-3p mimic also rescued the MT1JP overexpression-induced inhibitory effect on invasion in MDA-MB-231 cells (Figure 6C). In addition, the MT1JP overexpression-induced DDP sensitivity of DDP-resistant MDA-MB-231 cells was also rescued by miR-24-3p mimic (Figure 6D). These data indicate that the tumor suppression and cisplatin-sensitizing functions of MT1JP in breast cancer cells involves negative regulation of miR-24-3p.
Figure 6.
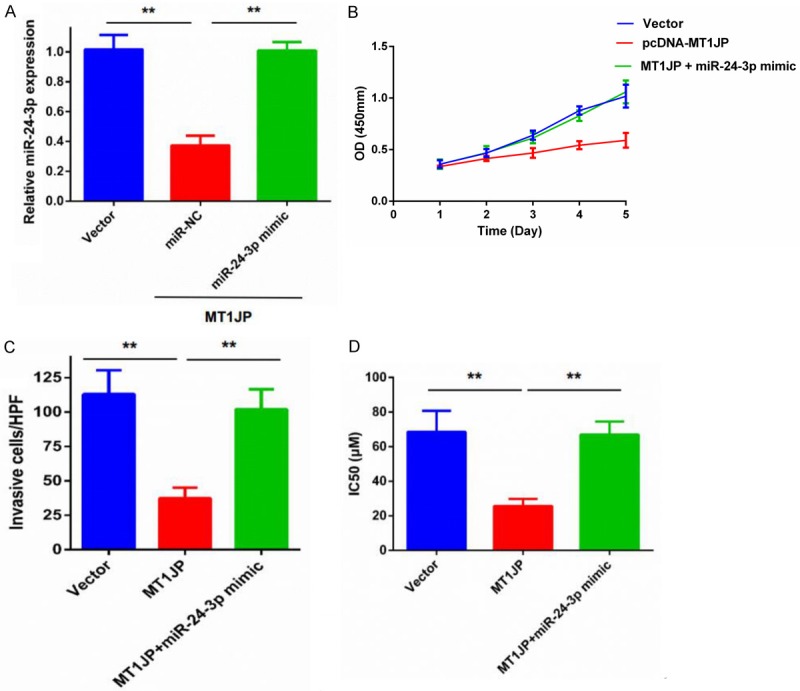
Tumor suppression and cisplatin sensitizing in breast cancer cells caused by MT1JP was partially reversed by co-transfection with a miR-24-3p mimic. A. qRT-PCR to measure miR-24-3p expression in MDA-MB-231 cells that were stably co-transfected with pMT1JP and miR-24-3p mimic or NC. ** P < 0.01. B. CCK-8 assays to measure proliferation of MDA-MB-231 cells that were stably transfected with pMT1JP and miR-24-3p mimic or NC. C. Transwell assays to measure migration and invasion of MDA-MB-231 cells that were stably transfected with pMT1JP and miR-24-3p mimic or NC. ** P < 0.01. D. Cisplatin sensitivity of MDA-MB-231DDP cells that were stably transfected with pMT1JP and miR-24-3p mimic or NC. ** P < 0.01. NC: Negative control.
MT1JP overexpression affected the Wnt/β-catenin signaling pathway
We performed western blot analysis to investigate whether MT1JP overexpression affected the Wnt/β-catenin signaling pathway. The results showed that MT1JP overexpression significantly inhibited β-catenin expression compared to negative control groups. Furthermore, we found that MT1JP overexpression significantly suppressed the expression of cyclin D1 and c-myc, which are important downstream genes of the Wnt/β-catenin signaling pathway (Figure 7).
Figure 7.
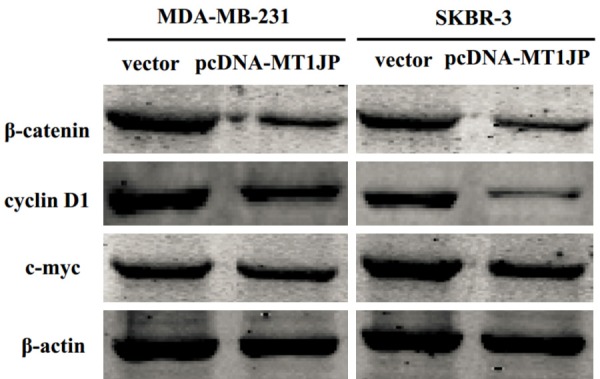
MT1JP overexpression affected the Wnt/β-catenin signaling pathway. Western blot analysis determining effect of MT1JP overexpression on the expression level of β-catenin, cyclin D1, and c-myc expression in the MDA-MB-231 and SKBR-3 cells.
Discussion
Mounting evidence suggests that lncRNAs are important to the regulation of cell differentiation, proliferation, and apoptosis [16,17]. It is also known that aberrant lncRNA has crucial roles in the initiation and development of various human cancers, including breast cancer. For example, Liu et al. reported that MT1JP suppresses tumor progression via a p53-related signaling pathway [12]. Another report found that MT1JP can competitively bind endogenous miR-214-3p and regulate RUNX3 expression, further supporting involvement in tumorigenesis and progression of gastric cancer [14]. In addition, MT1JP also regulates the progression of gastric cancer by functioning as a competing endogenous RNA that competitively binds to miR-92a-3p and regulates FBXW7 expression [13].
In this study, we found that MT1JP expression was down-regulated in breast cancer tissues and cell lines. The down-regulated MT1JP expression was associated with tumor size and TNM stage. More importantly, breast cancer patients with higher MT1JP expression in cancer tissues had significantly better prognoses than breast cancer patients with lower MT1JP expression in cancer tissues. We also found that MT1JP overexpression significantly inhibited proliferation, colony formation, and invasion, and enhanced the cisplatin sensitivity of breast cancer cells in vitro. These data indicated that MT1JP was significantly correlated with the progression and prognosis of breast cancer patients and could serve as a novel biomarker for breast cancer diagnosis. However, our results showed that MT1JP acted as a tumor suppressor in breast cancer, which is consistent with previous studies that reported tumor-suppressing functions for MT1JP in gastric cancer.
Here we present the first evidence, to the best of our knowledge, that MT1JP can enhance DDP sensitivity in DDP-resistant breast cancer cells. DDP represents the first generation of platinum drugs and is an effective cell cycle nonspecific anti-cancer drug [18]. It has been widely used to treat a number of solid malignancies, including breast cancer [19,20]. DDP’s anti-cancer roles are mainly mediated by binding to DNA nucleobases and inducing DNA damage to trigger apoptosis in cancer cells [21,22]. Although breast cancer patients often have good initial responses to DDP-based chemotherapy, DDP resistance usually occurs. Therefore, enhancing DDP sensitivity in DDP-resistant breast cancer cells is extremely important for the treatment of breast cancer. Our results suggest that MT1JP may provide a novel target to combat DDP resistance and could serve as a biological indicator of DDP sensitivity in breast cancer treatment.
Recently, many studies have suggested that novel regulatory mechanisms in cancer development involve interactions between lncRNAs and miRNAs [23,24]. For example, lncRNA can regulate miRNAs by acting as molecular sponges or ceRNA [25]. One study found that lncRNA MALAT1 promoted growth and metastasis of bladder transitional cell carcinoma by targeting miR-124 [26]. Zhang et al. reported that lncRNA UCA1 promotes prostate cancer progression by acting as a ceRNA of AFT2 [27]. In this study, we found that MT1JP binds miR-24-3p, which is a oncogene in serval human cancer, such as lung cancer [28], renal cell carcinoma [29], and bladder cancer [30]. We performed rescue experiments to investigate whether miR-24-3p reversed the biological effects of MT1JP in breast cancer, finding that MT1JP inhibited breast cancer cell proliferation and invasion, and also enhanced DDP sensitivity. However, co-transfection with miR-24-3p mimic partially reversed these biological effects, suggesting that MT1JP effects on breast cancer cell are mediated by repressing miR-24-3p.
Abnormal activation of the Wnt/β-catenin signaling pathway is widely recognized as an important mechanism for breast cancer initiation and metastasis [31]. The Wnt/β-catenin signaling pathway is involved in maintaining cancer stem cell properties, and plays a critical role in the epithelial-mesenchymal transition (EMT) [32]. To further explore the possible molecular mechanisms for the tumor inhibiting effect of MT1JP in breast cancer, we performed western blots to determine the effect of MT1JP on the Wnt/β-catenin signaling pathway. Our results showed that MT1JP overexpression significantly inhibited the expression of β-catenin, cyclin D1, and c-myc in breast cancer cells. This finding indicates that MT1JP overexpression inhibited the Wnt/β-catenin signaling pathway.
Taken together, this is the first finding of decreased expression of MT1JP in breast cancer tissues and cell lines. Furthermore, we found that MT1JP inhibited breast cancer cell proliferation and invasion, and also enhanced DDP sensitivity in breast cancer cells. Our data indicate that the mechanism for these effects is MT1JP binding and repressing miR-24-3p, thereby inhibiting the Wnt/β-catenin signaling pathway. This study provides a novel insight that could aid the development of potential prognostic indicators for breast cancer patients and improve the therapeutic effects of cisplatin in breast cancer treatment.
Acknowledgements
This research was supported in part by the National Nature Science Foundation of China (81573008), and the Fund of Pudong Health Bureau of Shanghai (PWRd2014-01).
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Lu L, Dong J, Wang L, Xia Q, Zhang D, Kim H, Yin T, Fan S, Shen Q. Activation of STAT3 and Bcl-2 and reduction of reactive oxygen species (ROS) promote radioresistance in breast cancer and overcome of radioresistance with niclosamide. Oncogene. 2018;37:5292–5304. doi: 10.1038/s41388-018-0340-y. [DOI] [PubMed] [Google Scholar]
- 3.Liu J, Song Z, Feng C, Lu Y, Zhou Y, Lin Y, Dong C. The long non-coding RNA SUMO1P3 facilitates breast cancer progression by negatively regulating miR-320a. Am J Transl Res. 2017;9:5594–5602. [PMC free article] [PubMed] [Google Scholar]
- 4.Jastrzebski K, Thijssen B, Kluin RJ, de Lint K, Majewski IJ, Beijersbergen RL, Wessels LFA. Integrative modeling identifies key determinants of inhibitor sensitivity in breast cancer cell lines. Cancer Res. 2018;78:4396–4410. doi: 10.1158/0008-5472.CAN-17-2698. [DOI] [PubMed] [Google Scholar]
- 5.Ulker D, Ersoy YE, Gucin Z, Muslumanoglu M, Buyru N. Downregulation of SCARA5 may contribute to breast cancer via promoter hypermethylation. Gene. 2018;673:102–106. doi: 10.1016/j.gene.2018.06.036. [DOI] [PubMed] [Google Scholar]
- 6.Liu X, Bi L, Wang Q, Wen M, Li C, Ren Y, Jiao Q, Mao JH, Wang C, Wei G, Wang Y. miR-1204 targets VDR to promotes epithelial-mesenchymal transition and metastasis in breast cancer. Oncogene. 2018;37:3426–3439. doi: 10.1038/s41388-018-0215-2. [DOI] [PubMed] [Google Scholar]
- 7.Burki TK. Responses to carboplatin in BRCA1/2-mutated breast cancer. Lancet Oncol. 2018;19:e337. doi: 10.1016/S1470-2045(18)30407-8. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Tao Y, Liao Q. Long noncoding RNA: a crosslink in biological regulatory network. Brief Bioinform. 2018;19:930–945. doi: 10.1093/bib/bbx042. [DOI] [PubMed] [Google Scholar]
- 9.Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, Jiang Z, Xu J, Liu Q, Cao X. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344:310–3. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- 10.Wang X, Vukovic L, Koh HR, Schulten K, Myong S. Dynamic profiling of double-stranded RNA binding proteins. Nucleic Acids Res. 2015;43:7566–7576. doi: 10.1093/nar/gkv726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hua F, Li K, Yu JJ, Lv XX, Yan J, Zhang XW, Sun W, Lin H, Shang S, Wang F, Cui B, Mu R, Huang B, Jiang JD, Hu ZW. TRB3 links insulin/IGF to tumour promotion by interacting with p62 and impeding autophagic/proteasomal degradations. Nat Commun. 2015;6:7951. doi: 10.1038/ncomms8951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu L, Yue H, Liu Q, Yuan J, Li J, Wei G, Chen X, Lu Y, Guo M, Luo J, Chen R. LncRNA MT1JP functions as a tumor suppressor by interacting with TIAR to modulate the p53 pathway. Oncotarget. 2016;7:15787–15800. doi: 10.18632/oncotarget.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang G, Li S, Lu J, Ge Y, Wang Q, Ma G, Zhao Q, Wu D, Gong W, Du M, Chu H, Wang M, Zhang A, Zhang Z. LncRNA MT1JP functions as a ceRNA in regulating FBXW7 through competitively binding to miR-92a-3p in gastric cancer. Mol Cancer. 2018;17:87. doi: 10.1186/s12943-018-0829-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Y, Zhang G, Zou C, Zhang H, Gong Z, Wang W, Ma G, Jiang P, Zhang W. LncRNA MT1JP suppresses gastric cancer cell proliferation and migration through MT1JP/MiR-214-3p/RUNX3 axis. Cell Physiol Biochem. 2018;46:2445–2459. doi: 10.1159/000489651. [DOI] [PubMed] [Google Scholar]
- 15.Song Z, Feng C, Lu Y, Lin Y, Dong C. PHGDH is an independent prognosis marker and contributes cell proliferation, migration and invasion in human pancreatic cancer. Gene. 2018;642:43–50. doi: 10.1016/j.gene.2017.11.014. [DOI] [PubMed] [Google Scholar]
- 16.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilmes A, Bielow C, Ranninger C, Bellwon P, Aschauer L, Limonciel A, Chassaigne H, Kristl T, Aiche S, Huber CG, Guillou C, Hewitt P, Leonard MO, Dekant W, Bois F, Jennings P. Mechanism of cisplatin proximal tubule toxicity revealed by integrating transcriptomics, proteomics, metabolomics and biokinetics. Toxicol In Vitro. 2015;30:117–127. doi: 10.1016/j.tiv.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 19.Galluzzi L, Senovilla L, Vitale I, Michels J, Martins I, Kepp O, Castedo M, Kroemer G. Molecular mechanisms of cisplatin resistance. Oncogene. 2012;31:1869–1883. doi: 10.1038/onc.2011.384. [DOI] [PubMed] [Google Scholar]
- 20.Armstrong DK, Bundy B, Wenzel L, Huang HQ, Baergen R, Lele S, Copeland LJ, Walker JL, Burger RA Gynecologic Oncology Group. Intraperitoneal cisplatin and paclitaxel in ovarian cancer. N Engl J Med. 2006;354:34–43. doi: 10.1056/NEJMoa052985. [DOI] [PubMed] [Google Scholar]
- 21.O’Grady S, Finn SP, Cuffe S, Richard DJ, O’Byrne KJ, Barr MP. The role of DNA repair pathways in cisplatin resistant lung cancer. Cancer Treat Rev. 2014;40:1161–1170. doi: 10.1016/j.ctrv.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 22.Wu G, Zhou W, Pan X, Sun Y, Xu H, Shi P, Li J, Gao L, Tian X. miR-100 reverses cisplatin resistance in breast cancer by suppressing HAX-1. Cell Physiol Biochem. 2018;47:2077–2087. doi: 10.1159/000491476. [DOI] [PubMed] [Google Scholar]
- 23.Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17:272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 24.Ding J, Yeh CR, Sun Y, Lin C, Chou J, Ou Z, Chang C, Qi J, Yeh S. Estrogen receptor beta promotes renal cell carcinoma progression via regulating LncRNA HOTAIR-miR-138/200c/204/217 associated CeRNA network. Oncogene. 2018;37:5037–5053. doi: 10.1038/s41388-018-0175-6. [DOI] [PubMed] [Google Scholar]
- 25.Chen X, Chen Z, Yu S, Nie F, Yan S, Ma P, Chen Q, Wei C, Fu H, Xu T, Ren S, Sun M, Wang Z. Long noncoding RNA LINC01234 functions as a competing endogenous RNA to regulate CBFB expression by sponging miR-204-5p in gastric cancer. Clin Cancer Res. 2018;24:2002–2014. doi: 10.1158/1078-0432.CCR-17-2376. [DOI] [PubMed] [Google Scholar]
- 26.Jiao D, Li Z, Zhu M, Wang Y, Wu G, Han X. LncRNA MALAT1 promotes tumor growth and metastasis by targeting miR-124/foxq1 in bladder transitional cell carcinoma (BTCC) Am J Cancer Res. 2018;8:748–760. [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang S, Dong X, Ji T, Chen G, Shan L. Long non-coding RNA UCA1 promotes cell progression by acting as a competing endogenous RNA of ATF2 in prostate cancer. Am J Transl Res. 2017;9:366–375. [PMC free article] [PubMed] [Google Scholar]
- 28.Yan L, Ma J, Zhu Y, Zan J, Wang Z, Ling L, Li Q, Lv J, Qi S, Cao Y, Liu Y, Cao L, Zhang Y, Qi Z, Nie L. miR-24-3p promotes cell migration and proliferation in lung cancer by targeting SOX7. J Cell Biochem. 2018;119:3989–3998. doi: 10.1002/jcb.26553. [DOI] [PubMed] [Google Scholar]
- 29.Jin L, Li Y, Nie L, He T, Hu J, Liu J, Chen M, Shi M, Jiang Z, Gui Y, Yang S, Lai Y. MicroRNA242 is associated with cell proliferation, invasion, migration and apoptosis in renal cell carcinoma. Mol Med Rep. 2017;16:9157–9164. doi: 10.3892/mmr.2017.7705. [DOI] [PubMed] [Google Scholar]
- 30.Yu G, Jia Z, Dou Z. miR-24-3p regulates bladder cancer cell proliferation, migration, invasion and autophagy by targeting DEDD. Oncol Rep. 2017;37:1123–1131. doi: 10.3892/or.2016.5326. [DOI] [PubMed] [Google Scholar]
- 31.Li Y, Jin K, van Pelt GW, van Dam H, Yu X, Mesker WE, Ten Dijke P, Zhou F, Zhang L. c-Myb enhances breast cancer invasion and metastasis through the Wnt/beta-catenin/Axin2 pathway. Cancer Res. 2016;76:3364–3375. doi: 10.1158/0008-5472.CAN-15-2302. [DOI] [PubMed] [Google Scholar]
- 32.Ghahhari NM, Babashah S. Interplay between microRNAs and WNT/beta-catenin signalling pathway regulates epithelial-mesenchymal transition in cancer. Eur J Cancer. 2015;51:1638–1649. doi: 10.1016/j.ejca.2015.04.021. [DOI] [PubMed] [Google Scholar]


