Fig. 1.
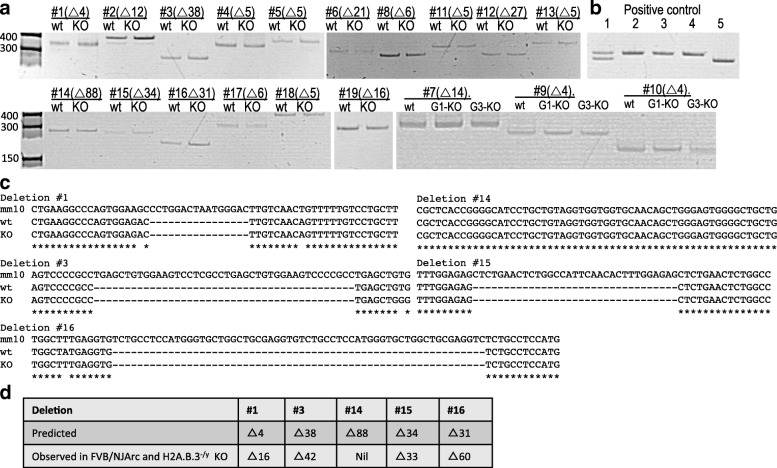
Interrogation of predicted off-target heterozygous deletions in H2A.B.3−/y mice. a 300–400-bp regions surrounding 19 common predicted heterozygous deletions were amplified from gDNA of H2A.B.3−/y(KO) and FVB/NJArc in-house wt mice. PCR product sizes for wt and H2A.B.3−/y were compared by resolving the products on a 7% polyacrylamide gel, side by side. The predicted deletion is indicated with △, followed by the number of nucleotides (nt) predicted to be deleted. b Positive controls showing that a deletion as small as 5 and 10 nt can be resolved on 7% polyacrylamide gel. Lane 1, a heterozygous △5 nt, lanes 2, 3, 4 wt.; lane 5, homozygous △10 nt. c 5 out of the 19 PCR products were sequenced by Sanger sequencing to show that FVB/NJArc and H2A.B.3−/y mice have identical sequences but they are different from the mm10 reference genome. d Table showing that the prediction tools are often inaccurate at predicting the presence or the size of a putative deletion