Figure 1. Summary for functional and structural identification of unknown metabolites.
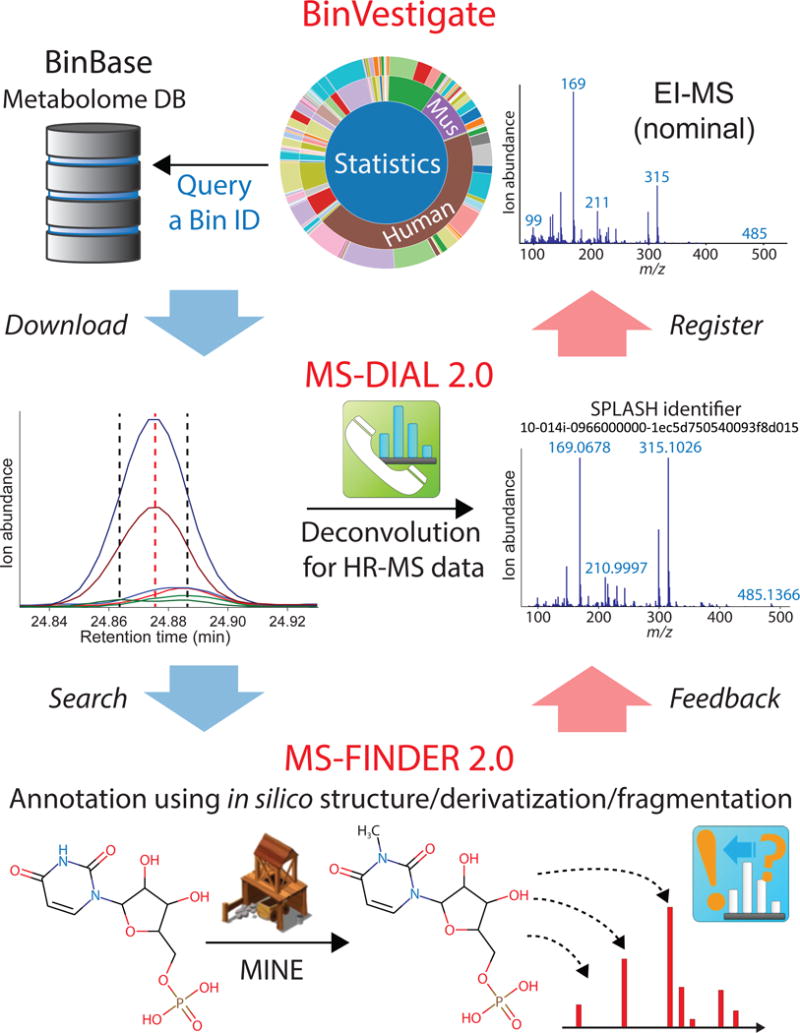
(a) BinVestigate to search unknown compounds for metabolomics study metadata and (nominal) EI-MS spectra in BinBase, with results shown as sunburst diagrams to illustrate the biological origin (species, organs, cell types) of unknowns. (b) MS-DIAL 2.0 for universal GC-MS or LC-MS/MS deconvolution with high resolution (HR) mass spectrometry analytics to obtain the deconvoluted HR-MS spectra of unknowns needed for compound identification. (c) MS-FINDER 2.0 for universal GC-EI-MS and LC-ESI-MS/MS spectral interpretation to annotate unknowns in combination with the enzyme promiscuity structure database (MINE), resulted in the discovery of biologically significant chemical structure. The tools are fully connected in MS-DIAL. Each tool is also available as standalone program.
