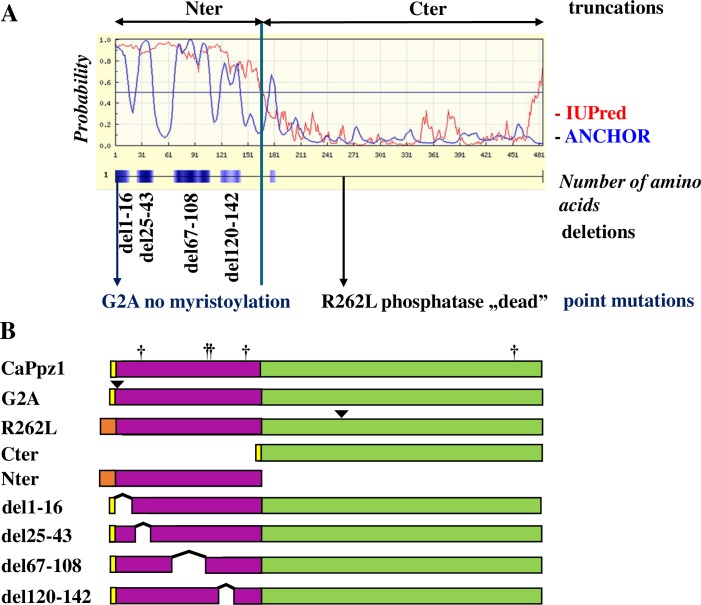
Fig 1. The rationale behind the in vitro mutagenesis of the CaPpz1 phosphatase.
A. Bioinformatic analysis of the CaPpz1 protein. The IUPred (red line) and ANCHOR (blue line) software revealed the disordered regions (above 0.5 probability) and four main protein binding sites (blue boxes) in the N-terminal domain of CaPpz1. In the four CaPpz1 deletion mutants, these potential binding sites were eliminated. In addition, two point mutants (the not myristoylated G2A and the inactive R262L) were generated, and the two main domains of the protein (Nter and Cter) were expressed separately. B. Schematic representation of the bacterially expressed recombinant proteins. The yellow boxes show the residual part of the GST-tag that remains in the bacterially expressed proteins after Prescission protease cleavage, while the light brown boxes represent the 6xHis tag of the recombinant proteins (note that these features are not present in the proteins expressed in S. cerevisiae). The N-terminal domain is violet and the C-terminal domain is green. Point mutations are labeled with a triangle, and five unintentional S to L exchanges due to the special codon usage of C. albicans are indicated by crosses.