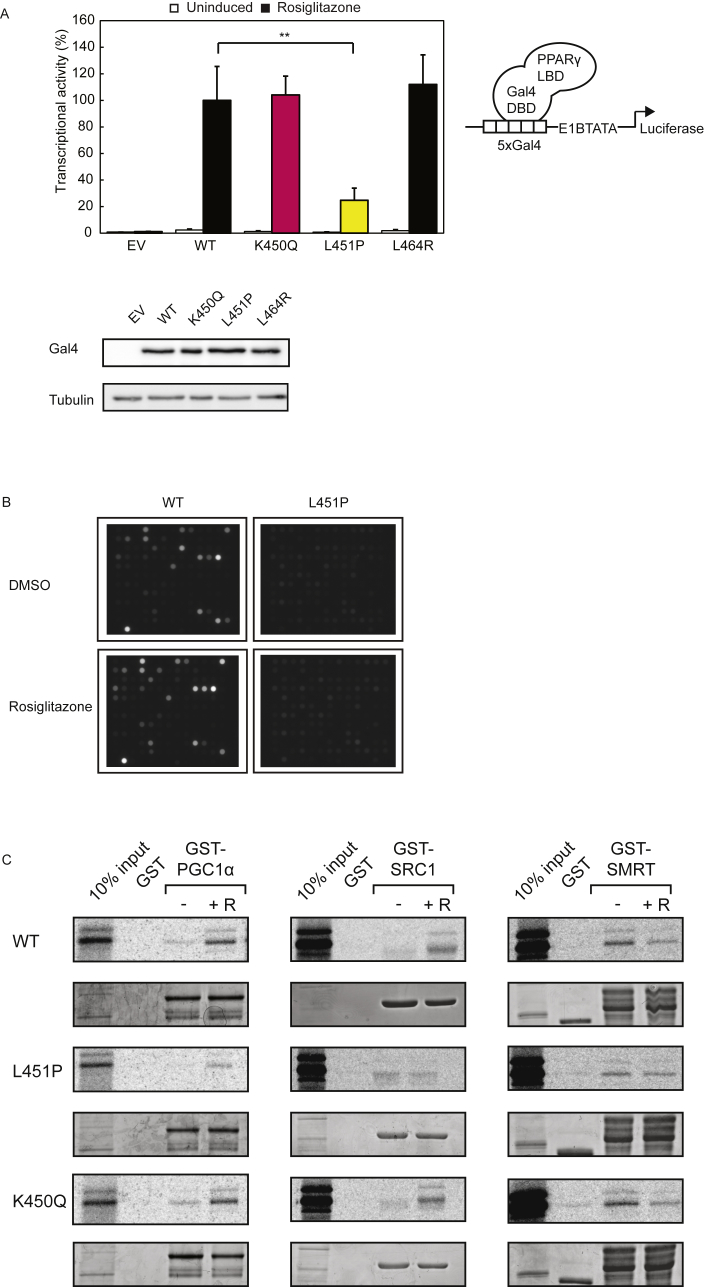
Figure 4.
PPARγ L451P mutant displays general ligand-mediated cofactor binding defects. A. U2OS cells were transiently transfected with chimeric Gal4 DBD-hPPARγ LBD wildtype and mutant fusion proteins and 5xGal4-E1BTATA-Luciferase. Cells were treated with or without 1 μM rosiglitazone. Shown results are average of three independent experiments assayed in duplicate ± SEM. **P < 0.01 cells transfected with mutant vs. WT. DBD, DNA binding domain; LBD, ligand binding domain. Comparable amounts of Gal4 DBD-hPPARγ LBD proteins were detected by western blot analysis using an antibody against Gal4 DBD. B. Pamgene® chips containing 154 different cofactor derived peptides (containing either LxxLL or LxxxIxxxL motifs) were incubated with recombinant GST-PPARγ-LBD or GST-PPARγ L451P-LBD and anti-GST-alexa, in the absence or presence of rosiglitazone. After 102 pump cycles a CCD camera recorded fluorescence (100 ms). Experiment was performed in triplo. Four representative images are shown. C. GST fusion proteins as indicated in the figure coupled to glutathione-Sepharose beads were incubated with [35S] methionine-labeled PPARγ (wildtype or mutant) in absence and presence of rosiglitazone (indicated by + R, 10 μM) to determine the effect of L451P on interactions with coregulators. GST alone was used as a negative control. 10% of the total lysate of the [35S] methionine-labeled PPARγ proteins used for the pull down assay was applied as control (input). Levels of GST-proteins have been confirmed by Coomassie staining.