Abstract
The α7 nicotinic acetylcholine receptor (α7nAChR) has been a promising target for diseases affecting cognition and higher cortical functions; however, the effect observed in animal models failed to translate into human clinical trials identifying a translational gap. CHRFAM7A is a human-specific fusion gene with properties that enable incorporation into the α7nAChR and, being human specific, CHRFAM7A effect was not accounted for in preclinical studies. We hypothesized that CHRFAM7A may account for this translational gap and understanding its function may offer novel insights when exploring α7nAChR as a drug target. CHRFAM7A is present in different copy number variations (CNV) in the human genome with high frequency. To study the functional consequences of the presence of the CHRFAM7A, two induced pluripotent stem cell (iPSC) lines (0 copy and 1 copy direct) were developed. The 0 copy line was rescued with CHRFAM7A transfection to control for genetic heterogeneity. As readouts for genotype–phenotype correlation, α7nAChR synaptic transmission and amyloid beta 1–42 (Aβ1–42) uptake were tested. Synaptic transmission in the presence of CHRFAM7A demonstrated that PNU-modulated desensitization of α7nAChR currents increased as a function of CHRFAM7A dosage. CHRFAM7A mitigated the dose response of Aβ1–42 uptake suggesting a protective effect beyond physiological concentrations. Furthermore, in the presence of CHRFAM7A Aβ1–42 uptake activated neuronal interleukin 1β (IL-1β) and tumor necrosis factor α (TNF-α) without activating the canonical inflammasome pathway. Lead optimization may identify more potent molecules when the screen has a model harboring CHRFAM7A. Incorporating pharmacogenetics into clinical trials may enhance signals in efficacy measures.
Introduction
The α7 nicotinic acetylcholine receptor (α7nAChR) is a ligand-gated ion channel implicated in cognition and neuropsychiatric disorders, including schizophrenia1–3, Alzheimer’s disease (AD)4,5 attention deficit hyperactivity disorder6, addiction7, pain8,9, and Parkinson disease10. Agonists and positive allosteric modulators (PAMs) of α7nAChR are being tested in clinical trials for central nervous system indications11, and a translational gap emerged12. While animal studies consistently demonstrate a cognitive benefit, this effect was not apparent in human trials13,14.
The α7nAChR is a homopentamer characterized by unique functional properties, including fast activation and desensitization by agonists, high Ca2+ permeability, and selective inhibition by α-bungarotoxin (α-BGT) and methyllycaconitine (MLA)12,15. It is expressed in brain regions underlying cognition and memory16, including the basal forebrain (nucleus basalis), hippocampus, neocortex, and amygdala17,18, α7nAChRs are expressed in neurons, microglia and astrocytes19. The α7nAChRs are regulators of the cholinergic anti-inflammatory pathway20,21, and acetylcholine (ACh) produces a dose-dependent inhibition of interleukin 6 (IL-6), IL-1β, and tumor necrosis factor α (TNF-α) in human macrophages22. In neuronal cell culture, Aβ1–42 binds with high affinity to the α7nAChRs and the receptor facilitates internalization of Aβ1–42 through endocytosis23.
One of the unique features of the α7nAChR in the human context is the presence of the fusion gene, CHRFAM7A24,25, The ancestral allele lacking the fusion gene is present in about 1% of the population, while 99% of the human population harbors the fusion gene. Further complexity is added as the orientation of CHRFAM7A can be direct or inverted, and the gene itself can be present in 1, 2, or even 3 copies, allowing for homozygous and heterozygous combinations. The predicted proteins of the direct and inverted alleles differ due to a 2 bp deletion in the inverted sequence causing a frameshift during protein translation26.
CHRFAM7A harbors exons 5–10 of CHRNA7 (transmembrane and intracellular domain) and 5 new exons of the FAM7 sequence (extracellular domain) corresponding to a part-functional CHRNA7. Two independent copy number variation (CNV) genome-wide association studies (GWAS) reported an association between CHRFAM7A dosage and AD; lower copy number and lower expression levels of the fusion gene is associated with AD27–29, In contrast, in schizophrenia and bipolar disorder, upregulation of CHRFAM7A was observed in the brain3 and association studies suggest a correlation with the inverted orientation (2 bp deletion)26. Despite its widespread implication in neuropsychiatric diseases, functional studies are sparse. In CHRFAM7A-transfected Xenopus oocytes, CHRFAM7A is a stoichiometric dominant-negative regulator of α7nAChR30,31.
Owing to the high frequency and complexity of the CHRFAM7A CNV in the human population, understanding its functional impact is imperative for interpreting α7nAChR targeting clinical trials. To study the functional consequences of the presence of the CHRFAM7A gene product on α7nAChR, we developed two induced pluripotent stem cell (iPSC) lines from skin biopsies of subjects affected by AD. UB068 has two ancestral haplotypes, thus it is lacking the CHRFAM7A gene (0 copy). UB052 has the CHRFAM7A direct orientation haplotype on one allele and the ancestral haplotype on the other allele (1 copy). iPSCs differentiated into relevant cell types, in our case medial ganglionic eminence (MGE) progenitors and neurons, model the effect of CHRFAM7A on α7nAChR function in the human context. As readouts for genotype–phenotype correlation, α7nAChR synaptic transmission and Aβ1–42 uptake were tested.
Materials and methods
Ethical statement, skin biopsy, and genotyping
The Institutional Review Board approved the study. The informed consents were obtained from the donors. Subjects requiring legally authorized representatives were excluded from the study.
iPSC generation and cell culture
iPSC lines (UB068—0 copy and UB052—1 copy direct) were generated from human skin biopsies in WNYSTEM (University at Buffalo) by episomal transformation and propagated in standard media (Supplementary data, Methods). iPSCs characterization according the industry standards included morphological assessment and live staining with the TRA-1–60 Alexa Fluor 488 Conjugate Kit (Life Technologies). Gene and protein expression for pluripotency/self-renewal and the three germ layer markers at gene and protein levels was assessed by reverse transcription–quantitative polymerase chain reaction (RT–qPCR) and immunocytochemistry (ICC) (Supplementary data, Methods; The primers (IDT) are listed in Supplementary data, Table 1, The primary antibodies are listed in Supplementary data, Table 2). Array comparative genome hybridization (aCGH) was performed between the iPSC colony and the original blood DNA sample from the same individual according to the manufacturer’s protocol. The ADM2 algorithm with a threshold of 6 was used to detect de novo events.
Pluripotency of the iPSC was confirmed by the TaqMan hPSC Scorecard Assay (Life Technologies) according to the manufacturer’s protocol32,33.
Neuronal differentiation and transfection
Neuronal differentiation of iPSC toward MGE progenitors and neurons was carried out using the protocol based on Liu et al.34 with modifications (Supplementary data, Methods). MGE progenitors were transfected with either pcDNA3.1-CHRFAM7A-mCherry (a gift from Henry Lester (Addgene plasmid # 62635)35) or with pcDNA3.3-mCherry (a gift from Derrick Rossi (Addgene plasmid # 26823)36 constructs according to Ma et al.37.
Total cell lysate preparation and immunoblotting
Total cell lysates from MGE progenitors were prepared using RIPA buffer (Cell Signaling Technologies) according to the manufacturer’s protocol. Twenty-five μg of total protein was separated on 4–20% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (Bio-Rad), transferred onto polyvinylidene difluoride membrane (Bio-Rad), and incubated overnight at 4 °C with primary antibodies (Supplementary data, Table 2). Specific immunoreactive bands were detected using ChemiDoc XRS + Imaging Systems (Bio-Rad).
α-BGT staining and confocal microscopy
Live α-BGT staining of neurons was carried out as described previously38. Briefly, neurons grown on 8-well glass chambers were pre-incubated for 10 min with 1 mM Nicotine followed by incubation for 30 min with 2 μM with α-BGT. Confocal images were captured by using LSM510 Meta microscope (×40 objective). Images were acquired using the ZEN black software (Zeiss).
HEK 293 cell culture and transfection
Human embryonic kidney (HEK) 293 cells were maintained in Dulbecco’s Minimal Essential Medium supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin, pH 7.4. Human α7nAChRs and CHRFAM7A were expressed in HEK 293 cells by transient transfection (CaPO4 precipitation method) of these cDNA in ratios of 4:1 or 1:4. To aid surface expression of α7, we co-transfected intracellular chaperones Ric-339 and NACHO40 in 1:1:1 ratio in all experiments.
Electrophysiology
Whole-cell and single-channel currents were recorded in the cell-attached patch configuration as described previously41 (Supplementary data, Methods). Kinetic analyses of single-channel currents were performed by using QuB42. Single-channel currents were idealized by segmental k-means algorithm. nPo was estimated by dividing the cumulative open probability by the number of channels in the patch (maximum number of overlaps of open current levels in the data) as follows:
Amyloid beta uptake and quantification
Amyloid beta uptake was performed using fluorescently labeled Aβ1–42 (AnaSpec) as described previously43. Briefly, MGE progenitors grown on glass coverslips were treated with various concentrations of Fluorescin-Aβ1–42 (1, 10, 25, 50, 100, and 250 nM) for 18 h. Live images were taken using EVOS (Life Technologies) microscope (×40 objective). Confocal images were taken with the LSM510 Meta microscope (×40 objective). Amyloid beta uptake was quantified by using ImageJ (imagej.nih.gov) or by Flow cytometry using LSRII-Fortessa with FACS DIVA (BD Biosciences). The flow cytometry data were analyzed using the FlowJo software (https://www.flowjo.com/). Data are presented as the average of triplicates ± standard deviation (SD).
IL-1β enzyme-linked immunosorbent assay (ELISA)
Following transfection and amyloid beta uptake, total cell lysates were collected and stored at −80 °C. Concentration of IL-1β in the cell lysates was estimated using a human-specific high-sensitivity IL-1β ELISA Kit (Thermo Fisher) according to the manufacturer’s protocol.
Statistical analysis
Values are expressed as means ± SD or ±SEM, as indicated in figure legends. Statistical significance was determined by an unpaired Student’s t-test (two-tailed). P values <0.05 were deemed statistically significant.
Results
iPSC characterization
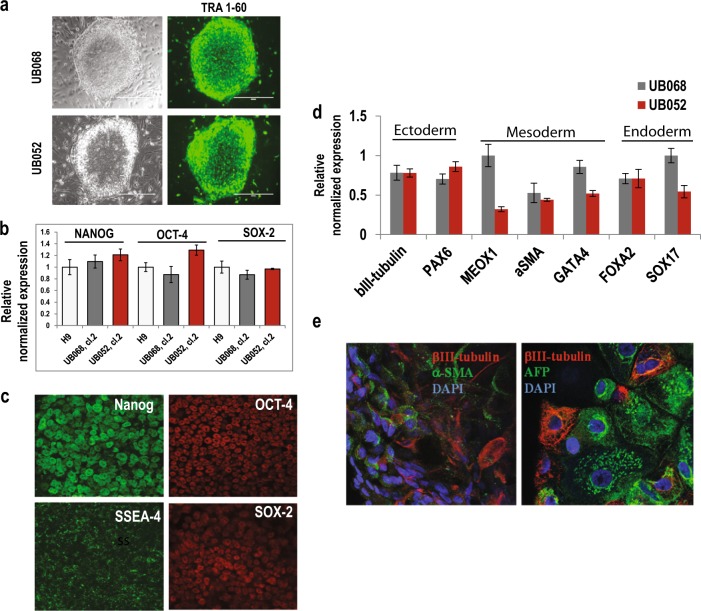
Stem cell characteristics of new iPSC lines UB068 (0 copy) and UB052 (1 copy) were confirmed by morphology; TRA1–60 live cell staining (Fig. 1a); detecting expression of pluripotency/self-renewal markers (NANOG, OCT-4, and SOX2) (Fig. 1b); and by ICC for Nanog, Oct-4, SOX2 and SSEA-4 (Fig. 1c). aCGH between the iPSC colony and the original blood DNA excluded de novo chromosomal aberrations and CNVs (Fig. 1a, Supplementary data). TaqMan assay specific to the breakpoint sequence confirmed CHRFAM7A CNV dosage (Fig. 1b, Supplementary data).
Fig. 1. Characterization of human induced pluripotent stem cell (iPSC) lines.
a Live images of the UB068 (0 copy) and UB052 (1 copy of CHRFAM7A) colonies stained with TRA1–60 antibody. b Reverse transcription–quantitative polymerase chain reaction analysis of self-renewal/pluripotency gene expression in iPSC lines in comparison with H9 cell line. c Confocal images of the iPSCs probed for Nanog, Oct4, Sox2, and SSEA-4 proteins. Gene (d) and protein (e) expression of the three germ layer markers at day 15 of non-directed differentiation in UB068 and UB052 lines
Pluripotency of iPSC lines was confirmed by embryoid body (EB)-based non-directed differentiation (Fig. 1c, Supplementary data), gene expression (Fig. 1d), and ICC with anti-βIII-tubulin (ectoderm), anti-α-SMA (mesoderm), and anti-α-fetoprotein (AFP, endoderm) antibodies (Fig. 1e) and by the TaqMan hPSC Scorecard Assay (ThermoFisher) (Fig. 1d, Supplementary data).
Neuronal differentiation
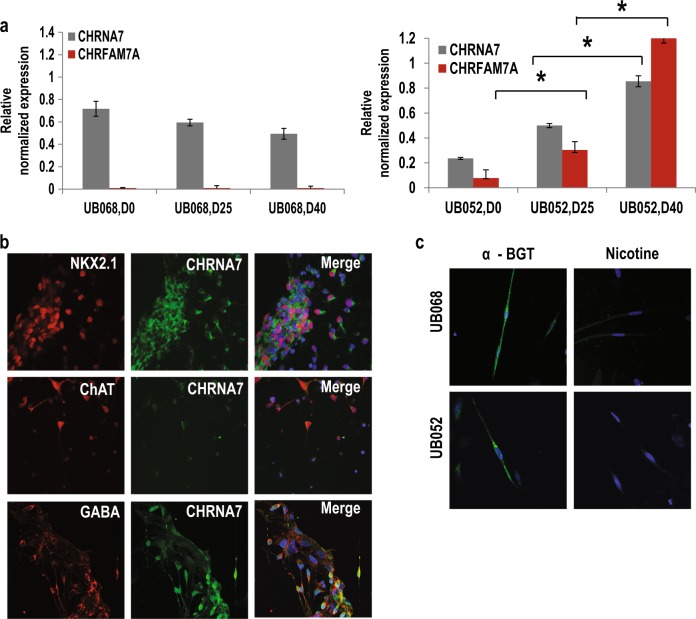
Based on the TaqMan hPSC Scorecard Assay clones with the highest preference for neuronal differentiation (UB068, clone 2 and UB052, clone 2) were chosen for further experiments. UB068 (0 copy) and UB052 (1 copy) clones were differentiated into MGE progenitors34 and MGE-derived BFCNs and GABA interneurons. Expression levels of NKX2.1, LHX6, and LHX8 (MGE markers), FOXG1 (forebrain marker), PAX6 (dorsal forebrain marker) SOX2 and MAP-2 (pan neuronal markers), and ChAT, GAD, TH, and HB9 (BFCN, GABA, dopaminergic, and motor neuron markers, respectively) were quantified during neuronal differentiation (Fig. 2a, Supplementary data). MGE progenitors demonstrated high level of FOXG1, NKX2.1, LHX6, and LHX8 expression detected at D25 and MGE-derived GABA and BFCN showed high level of GAD and ChAT expression and very low level of TH and HB9 expression at D40. ICC confirmed GABA and/or choline acetyltransferase (ChAT) expression in neurons (Fig. 2b, Supplementary data). Spontaneous action current activity and voltage-gated Na+ and K+ currents recorded from UB068- and UB052-derived neuronal cultures confirmed functional neurons (Fig. 2c, Supplementary data). Single-channel patch-clamp analysis confirmed α7-specific currents in neurons (Fig. 2d, Supplementary data).
Fig. 2. Expression of CHRNA7 and CHRFAM7A during neuronal differentiation of induced pluripotent stem cells (iPSCs).
a Reverse transcription–quantitative polymerase chain reaction analysis of CHRFAM7A and CHRNA expression throughout the course of neural differentiation: D0 (pluripotent iPSC), D25 (MGE progenitors), and D40 (neurons). Primers amplify the unique breakpoint sequence and distinguish between the CHRFAM7A and CHRNA7 mRNA. Data are presented as mean ± SEM. *P < 0.05 difference between CHRFAM7A expression at D0 and D25, and D40. b Confocal images demonstrating expression of CHRNA7/CHRFAM7A in MGE progenitors (NKX 2.1), GABA interneurons (GABA), and BFCN (choline acetyltransferase). Note that CHRNA7 and CHRMFAM7A cannot be distinguished with antibodies. c α7nAChR (α7 nicotinic acetylcholine receptor)-specific live staining of UB068- and UB052-derived neurons with α-bungarotoxin (α-BGT). Preincubation with nicotine prevents fluorescein isothiocyanate–α-BGT binding
CHRFAM7A gene expression during neuronal differentiation
CHRNA7 and CHRFAM7A breakpoint (unique sequence) specific primers demonstrated expression of CHRNA7 (in UB068 and UB052) and CHRFAM7A (in UB052) during neuronal differentiation. The results of RT–qPCR revealed a two-fold increase in CHRNA7 expression at D40 compared to D0 in UB052 cells (P = 0.045), while no changes in CHRNA7 expression were detected in UB068 cells (Fig. 2a, left and right panel). CHRFAM7A expression level was upregulated six times during the process of neuronal differentiation in UB052 (P = 0.027) [Fig. 2a, right panel]. ICC demonstrated co-expression of CHRNA7 and NKX2.1 in MGE progenitors (Fig. 2b) and MGE-derived neurons (both BFCN and GABA interneurons) generated from UB068 and UB052 lines (Fig. 2b). Live staining with α-BGT demonstrated the presence of functional α7nAChR in both UB068- and UB052-derived neurons (Fig. 2c).
CHRFAM7A is a modulator of the α7nAChR
The effect of CHRFAM7A on α7nAChRs can be attributed to altered biophysical properties and/or surface expression of the channels. To investigate the possible changes in the kinetic properties of α7nAChRs, single-channel cell-attached (mechanism of action) and whole-cell patch-clamp (surface expression) recordings on UB068 and UB052 neurons were performed.
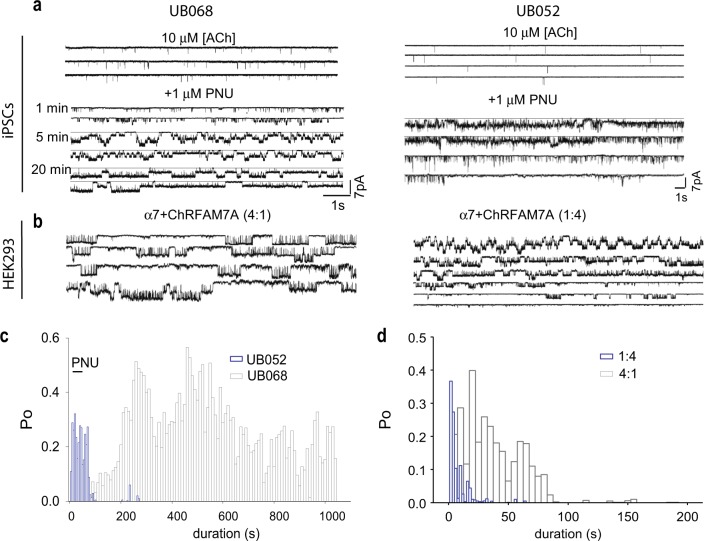
Functional expression of neuronal α7nAChRs was studied by recording cell-attached single-channel currents from UB068 (0 copy) and UB052 (1 copy) cell lines in the presence of TTX and TAE (Fig. 3a, left and right panels). α7nAChR expression in both cell types albeit lower expression in case of UB052 was observed. α7nAChRs currents were pharmacologically characterized by utilizing the PAM of α7nAChR, PNU 120596 (PNU). The effects of PNU on single α7nAChR currents from UB068 and UB052 cells are depicted in Fig. 3a, left and right panels. PNU in UB068 cells progressively increased channel open probability of single α7nAChRs in a time-dependent manner44,45, On the other hand, in UB052-derived neurons, PNU-modulated currents desensitized/ran down faster than in neurons generated from UB068. Comparative nPo analyses of currents from both cell types showed the differences in CHRFAM7A effects on the channel kinetic properties (Fig. 3c).
Fig. 3. Effect of CHRFAM7A on electrophysiological properties of α7 nicotinic acetylcholine receptors (α7nAChRs).
a Single-channel current recordings in cell-attached mode from neurons derived from UB068 (left panel) and UB052 (right panel) cell lines in the presence of TTX and TAE. Note the strong potentiation of α7 nAChRs currents in the presence of PNU. b Effect of CHRFAM7A on α7nAChR stoichiometry and functional properties in HEK 293 cells transfected with α7nAChR and CHRFAM7A cDNA in 4:1 (left panel) and 1:4 proportions (right panel); PNU differentially modulates currents from HEK 293 cells expressing α7nAChR and CHRFAM7A in 4:1 and 1:4 ratio. c Comparative nPo analyses of currents from both induced pluripotent stem cell lines showing difference in kinetic properties between the α7nAChR types expressed in them. d nPo analyses of both currents in HEK 293 cells transfected with the target genes
To probe the effect of CHRFAM7A on α7nAChR stoichiometry and functional properties, we heterologously expressed them by transfecting CHRNA7 and CHRFAM7A cDNA in HEK 293 cells in 4:1 and 1:4 proportions. The dosage of cDNA was expected to determine the stoichiometry of the α7nAChR expressed in HEK 293 cells. PNU modulated currents from HEK 293 cells expressing α7nAChR and CHRFAM7A both in 4:1 and 1:4 ratios (Fig. 3b, left and right panels respectively). Qualitatively, the single-channel current clusters appeared similar in both cases. However, nPo analyses of both currents suggest that desensitization of PNU-modulated α7nAChR currents increased as a function of CHRFAM7A dosage (Fig. 3d), consistent with the results contrasting UB068 and UB052.
Amyloid beta uptake via the α7nAChR is mitigated by CHRFAM7A
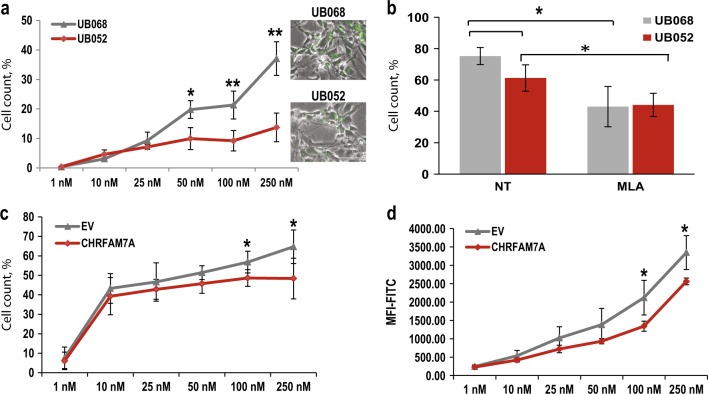
Aβ1–42 uptake in MGE progenitors derived from UB068 (0 copy), and UB052 (1 copy) lines was quantified by cell counts (live EVOS and confocal images, ImageJ) and flow cytometry. The results demonstrated higher Fluorescin-Aβ1–42 uptake in UB068 in contrast to UB052 (Fig. 4a). Furthermore, while the uptake was dose dependent (correlating with increasing concentration of Fluorescin-Aβ1–42 from 1 to 250 nM) in UB068, it was constant between concentrations of 25–250 nM in UB052 (Fig. 4a). This suggests that CHRFAM7A may be functioning as a regulator of Aβ1–42 uptake beyond physiological concentrations.
Fig. 4. CHRFAM7A modulates amyloid beta uptake via the α7 nicotinic acetylcholine receptor.
a Difference in concentration-dependent Aβ1–42 uptake by MGE progenitors derived from UB068 and UB052 lines. Data are presented as mean ± SD. *P < 0.05 **P < 0.001—difference between Aβ1–42 uptake in UB052 line compared to UB068 at each given Aβ1–42 concentration. Inset: representative EVOS images of Aβ1–42 (100 nM) uptake in both cell lines. b Pretreatment with α7-selective antagonist methyllycaconitine (MLA; 10 μM) leads to a significant decrease in Aβ1–42 (100 nM) uptake in both cell lines. Data are presented as mean ± SD. *P < 0.05—difference between Aβ1–42 uptake in MGE progenitors with and without treatment (NT) with MLA. Transfection of UB068 (0 copy) with CHRFAM7A causes a decrease in Aβ1–42 uptake in a concentration-dependent manner compared to transfection with empty vector (EV) analyzed by cell counts (c) and as mean fluorescent intensity by flow cytometry (d). Data are presented as mean ± SD. *P < 0.05—difference between Aβ1–42 uptake in CHRFAM7A-transfected cells compared to EV-transfected cells at each given Aβ1–42 concentration
Pharmacological modulation of the α7nAChR affected Aβ1–42 uptake suggesting an α7AChR-dependent mechanism. Pretreatment of MGE progenitors with the α7-selective antagonist MLA resulted in a significant decrease in Aβ1–42 uptake in neuronal progenitor cells (NPCs) derived from UB068 (from 74 to 43%; P = 0.009) and from UB052 (from 58 to 44%; P = 0.037) (Fig. 4b).
To exclude that the observed effect is caused by the different genetic background in the two lines, UB068-derived NPCs were transfected with pcDNA3.1-CHRFAM7A-mCherry cDNA (controls: pcDNA3.3-mCherry empty vector (EV)). Efficiency of transfection was estimated by counting mCherry-positive cells and confirmed by qPCR with specific primers (Fig. 3, Supplementary data). After 24 h of transfection, the cells were treated for 18 h with the concentration gradient of Fluorescin-Aβ1–42 (1–250 nM). Aβ1–42 uptake demonstrated that the presence of CHRFAM7A leads to a decrease in Aβ1–42 uptake (Fig. 4c). Furthermore, the regulatory effect of CHRFAM7A on Aβ1–42 uptake observed in UB052-derived NPCs between the concentrations from 25 to 250 nM was replicated. In contrast, transfection of the NPC with EV pcDNA3.3-mCherry resulted in a dose–response Aβ1–42 uptake similar to non-transfected UB068. Orthogonal quantification by flow cytometry corroborated these results (Fig. 4d and Fig. 4, Supplementary data).
Activation of inflammatory pathways by Aβ1–42 through the α7nAChR is mitigated by CHRFAM7A
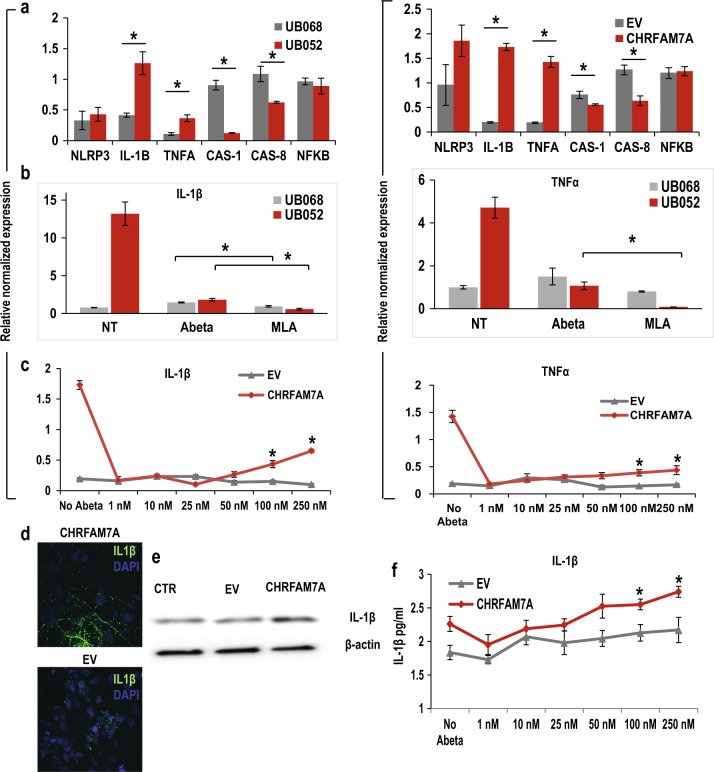
α7nAChRs are regulators of the cholinergic anti-inflammatory pathway and ACh produces a dose-dependent inhibition of IL-6, IL-1β, and TNF-α in macrophages. Therefore, we examined whether this mechanism plays a role in MGE progenitors expressing α7AChRs and whether it is modulated by the presence of CHRFAM7A. We hypothesized that the presence of CHRFAM7A affects the inflammatory pathways activated by Aβ1–42 uptake. We found that expression levels of IL-1B and TNFA are increased in MGE progenitors expressing CHRFAM7A (UB052 and UB068 transfected with CHRFAM7A; Fig. 5a—left and right panels), whereas CAS-1 and CAS-8 expression are higher in MGE progenitors lacking the fusion gene (UB068 and transfection with EV) suggesting that IL-1β activation in the presence of CHRFAM7A is not dependent on the canonical inflammasome pathway. ΝFΚΒ is not affected by the presence of CHRFAM7A (Fig. 5a—left and right panels). Pretreatment of NPCs with MLA (24 h) followed by Fluorescin-Aβ1–42 uptake (100 nM; 18 h) significantly decreased Aβ1–42-activated expression of IL-1B (Fig. 5b, left panel) and TNFA (Fig. 5b, right panel) in both lines suggesting an α7nAChR-dependent mechanism. Furthermore, Fluorescin-Aβ1–42 uptake increased expression of IL-1β and TNF-α in a concentration-dependent manner in CHRFAM7A-transfected cells but not in EV-transfected cells (Fig. 5c, left and right panels). The CHRFAM7A-dependent increase in IL-1β expression was validated by ICC (Fig. 5d) and immunoblotting (Fig. 5e). ELISA demonstrated that IL-1β is released from the cells and that the presence of CHRFAM7A causes a dose-dependent IL-1B release in response to Fluorescin-Aβ1–42 uptake (Fig. 5f).
Fig. 5. Activation of inflammatory pathways by Aβ1–42 through the α7 nicotinic acetylcholine receptor is mitigated by CHRFAM7A.
a Comparison of inflammasome-related gene expression profiles in neuronal progenitor cells (NPCs) generated from UB068 and UB052 (left panel) and in UB068 transfected with CHRFAM7A and/or empty vector (EV) (right panel). Data are presented as mean ± SEM. * - P < 0.05 - difference in gene expression levels in UB052 cells compared to UB068 and in CHRFAM7A-transfected cells compared to EV-transfected cells. b Pretreatment with α7-selective antagonist methyllycaconitine (MLA) decreases Aβ1–42-induced IL-1β (left panel) and tumor necrosis factor α (TNF-α; right panel) expression in UB068 and UB052 lines. 100 nM Aβ1–42 was used for treatment. Data are presented as mean ± SEM. *P < 0.05—difference in Aβ1–42-induced gene expression levels between MGE progenitors with and without treatment with MLA in both cell lines. c Expression of IL-1β (left panel) and TNF-α (right panel) correlates with Aβ1–42 uptake in concentration-dependent manner in the NPCs transfected with CHRFAM7A. Data are presented as mean ± SEM. *P < 0.05—difference in IL-1β and/or TNF-α expression in CHRFAM7A-transfected cells compared to EV-transfected cells at each given Aβ1–42 concentration. d Representative confocal images and e immunoblot analysis of total cell lysates showing an increase in IL-1β expression in the cells transfected with CHRFAM7A. f In MGE progenitors transfected with CHRFAM7A, Fluorescin-Aβ1–42 uptake induces a concentration-dependent increase in IL-1β secretion as detected by ELISA. Data are presented as mean ± SD. *P < 0.05—difference between IL-1 β concentration in CHRFAM7A-transfected cells compared to EV-transfected cells at each given Aβ1–42 concentration
Discussion
Human-specific genes are emerging culprits in complex human diseases46. CHRFAM7A, a human specific fusion gene in high frequency in the population, has been implicated in a broad array of neuropsychiatric disorders, including schizophrenia, bipolar disorder, dementia with Lewy bodies, Pick disease, and AD; all are human-specific diseases affecting association cortices and higher cognitive function25. While CHRN7A has been a promising target for diseases affecting cognition, the effect observed in animal models failed to translate in human clinical trials suggesting a human-specific mechanism13,14, we developed a model system to study the modifying effect of CHRFAM7A which (i) has the human biological context, (ii) allows studies in specific cell types, (iii) is a renewable source, and iv) is amenable to scaling. Adapting this iPSC system for high-throughput screening can advance drug discovery in diseases such as AD and schizophrenia and addresses unmet medical needs. In this work, we present two iPSC lines, one with the rare homozygous ancestral haplotype with no CHRFAM7A and another harboring a single direct copy of CHRFAM7A on one allele with the ancestral haplotype on the other. The CHRFAM7A CNV harboring iPSC line contrasted to the 0 copy state provides a unique experimental system, where its effect can be studied in disease-relevant differentiated cells. While the two lines are from two different individuals, transfection of the CHRFAM7A into the 0 copy line showed similar results when the transfection efficiency is taken into account, suggesting that the results are independent of the genetic background. There is a need for genome-edited isogenic lines, such as UB068 with inserted CHRFAM7A, and UB052 with knocking out CHRFAM7A to address CHRFAM7A effect on α7nAChR function.
iPSC lines underwent stringent characterization and selected colonies were successfully differentiated into MGE progenitors. CHRNA7 and CHRFAM7A expression increases with differentiation, and CHRFAM7A is preferentially induced when present in the cell line. Functional receptors demonstrated electrophysiological properties and response to pharmacological treatments with agonists, antagonists, and PAMs consistent with α7nAChR.
Interestingly, when CHRFAM7A was present either in the iPSC line UB052 or after transfection of HEK293 cells, electrophysiological properties of α7nAChR were different from the 0 copy line. PNU in UB068 cells (CHRFAM7A absent) progressively increased channel open probability of single α7nAChRs in a time-dependent manner44. On the other hand, in UB052, PNU-modulated currents ran down faster than in UB068. Comparative nPo analyses of currents from both cell types showed clear difference in kinetic properties whether CHRFAM7A was expressed or absent. When CHRNA7 and CHRFAM7A cDNA in HEK 293 cells were expressed in 4:1 and 1:4 proportions, PNU modulated currents differentially. Qualitatively, the single-channel current clusters appeared similar in both cases; however, nPo analyses of both currents suggest that desensitization of PNU-modulated α7nAChR currents increased as a function of CHRFAM7A dosage. These altered electrophysiological properties could suggest that, based on their genotype, individuals may respond differentially to PAMs and perhaps to agonists and antagonists as well. Thus, during drug development by targeting α7nAChR for indications such as cognition or negative symptoms of schizophrenia, taking into account these pharmacogenetic correlations may prove more powerful.
CHRFAM7A modified Aβ1–42 uptake mediated through the α7nAChR47–49,23, demonstrating a dose response to Aβ1–42 concentration and modulation of uptake by α7nAChR agonist, antagonists, and PAMs. Comparison of the CHRFAM7A null (0 copy) and heterozygous (1 copy) lines and transfection of CHRFAM7A into the null line demonstrated a regulatory effect of CHRFAM7A on Aβ1–42 uptake, which was concentration dependent. Linear uptake of Aβ1–42 was observed in the 0 line, while the presence of CHRFAM7A mitigated the dose response of Aβ1–42 uptake at higher concentrations, suggesting a protective effect beyond physiological concentrations. These observations align with the age-dependent penetrance of AD and the hypothesis of decreased clearance being the leading mechanism in sporadic AD50. We hypothesize that CHRFAM7A is protective in AD during the accumulation of Aβ1–42 by mitigating Aβ1–42 uptake and by activating neuronal IL-1β expression and release as cry for help. Since α7nAChR targeting clinical trials focused on synaptic modulation and cognitive benefit in the short term (12–14 weeks), its suggested disease-modifying effect has not been explored.
These results suggest a negative modulatory effect of CHRFAM7A on synaptic transmission (relevance in schizophrenia) and a modulatory effect on Aβ1–42 uptake (relevance in AD), consistent with the direction of the association signals in schizophrenia (increased CHRFAM7A as risk) and AD (loss of CHRFAM7A as risk). Since the CNV is frequent, lead optimization may identify more potent molecules when the screen has a model with CHRFAM7A. Pharmacogenetics needs to be implemented in the clinical trials as the presence of CHRFAM7A will likely have an impact on drug effect. Further studies are needed to evaluate whether there is a dosage effect of CHRFAM7A when present on both chromosomes, and even three copy individuals have been detected. Cell lines with the inverted copy of CHRFAM7A could answer the long lingering question whether the inverted copy is expressed and have a functional effect on the α7nAChR or if it behaves as a null allele. Isogenic lines can eliminate the genetic heterogeneity concern and are being developed. Using a genetically relevant model system and translating it into a genetically characterized population could result in markedly improved signal and is consistent with the efforts to deliver precision medicine.
Supplementary information
Acknowledgements
We thank Animesh Sinha, MD for performing the skin biopsies. We thank Jacqueline Heatwole for her technical assistance. This work was supported by Alzheimer Association AARG-16–443615, CTSA Pilot grant program, Edward A. and Stephanie E. Fial Fund, Community Foundation for Greater Buffalo, Dr. Louis Sklarow Memorial Trust and NIH grant GM121463.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41398-019-0375-z).
References
- 1.Freedman R, Hall M, Adler LE, Leonard S. Evidence in postmortem brain tissue for decreased numbers of hippocampal nicotinic receptors in schizophrenia. Biol. Psychiatry. 1995;38:22–33. doi: 10.1016/0006-3223(94)00252-X. [DOI] [PubMed] [Google Scholar]
- 2.Freedman R, et al. The genetics of sensory gating deficits in schizophrenia. Curr. Psychiatry Rep. 2003;5:155–161. doi: 10.1007/s11920-003-0032-2. [DOI] [PubMed] [Google Scholar]
- 3.Kunii Y, et al. CHRNA7 and CHRFAM7A mRNAs: co-localized and their expression levels altered in the postmortem dorsolateral prefrontal cortex in major psychiatric disorders. Am. J. Psychiatry. 2015;172:1122–1130. doi: 10.1176/appi.ajp.2015.14080978. [DOI] [PubMed] [Google Scholar]
- 4.Hoskin, J. L., Al-Hasan, Y. & Sabbagh, M. N. Nicotinic acetylcholine receptor agonists for the treatment of Alzheimer’s dementia: an update. Nicotine Tob. Res. 10.1093/ntr/nty116 (2018). [DOI] [PMC free article] [PubMed]
- 5.Parri HR, Hernandez CM, Dineley KT. Research update: Alpha7 nicotinic acetylcholine receptor mechanisms in Alzheimer’s disease. Biochem. Pharmacol. 2011;82:931–942. doi: 10.1016/j.bcp.2011.06.039. [DOI] [PubMed] [Google Scholar]
- 6.Wilens TE, Decker MW. Neuronal nicotinic receptor agonists for the treatment of attention-deficit/hyperactivity disorder: focus on cognition. Biochem. Pharmacol. 2007;74:1212–1223. doi: 10.1016/j.bcp.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma G, Vijayaraghavan S. Nicotinic receptors: role in addiction and other disorders of the brain. Subst. Abus. 2008;2008(1):81. [PMC free article] [PubMed] [Google Scholar]
- 8.Alsharari SD, Freitas K, Damaj MI. Functional role of alpha7 nicotinic receptor in chronic neuropathic and inflammatory pain: studies in transgenic mice. Biochem. Pharmacol. 2013;86:1201–1207. doi: 10.1016/j.bcp.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 9.Freitas K, Carroll FI, Damaj MI. The antinociceptive effects of nicotinic receptors alpha7-positive allosteric modulators in murine acute and tonic pain models. J. Pharmacol. Exp. Ther. 2013;344:264–275. doi: 10.1124/jpet.112.197871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quik M, Zhang D, McGregor M, Bordia T. Alpha7 nicotinic receptors as therapeutic targets for Parkinson’s disease. Biochem. Pharmacol. 2015;97:399–407. doi: 10.1016/j.bcp.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beinat C, Banister SD, Herrera M, Law V, Kassiou M. The therapeutic potential of α7 nicotinic acetylcholine receptor (α7 nAChR) agonists for the treatment of the cognitive deficits associated with schizophrenia. Cns. Drugs. 2015;29:529–542. doi: 10.1007/s40263-015-0260-0. [DOI] [PubMed] [Google Scholar]
- 12.Yang T, Xiao T, Sun Q, Wang K. The current agonists and positive allostericmodulators of à7 nAChR for CNS indications in clinical trials. Acta Pharm. Sin. 2017;B7:611–622. doi: 10.1016/j.apsb.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sadigh-Eteghad S, Mahmoudi J, Babri S, Talebi M. Effect of alpha-7 nicotinic acetylcholine receptor activation on beta-amyloid induced recognition memory impairment. Possible role of neurovascular function. Acta Cir. Bras. 2015;30:736–742. doi: 10.1590/S0102-865020150110000003. [DOI] [PubMed] [Google Scholar]
- 14.Lewis A, Schalkwyk G, Bloch M. Alpha-7 nicotinic agonists for cognitive deficits in neuropsychiatric disorders: a translational meta-analysis of rodent and human studies. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2017;75:45–53. doi: 10.1016/j.pnpbp.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zoli M, Pistillo F, Gotti C. Diversity of native nicotinic receptor subtypes in mammalian brain. Neuropharmacology. 2015;96(pt B):302–11. doi: 10.1016/j.neuropharm.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 16.Levin ED. alpha7-Nicotinic receptors and cognition. Curr. Drug Targets. 2012;13:602–606. doi: 10.2174/138945012800398937. [DOI] [PubMed] [Google Scholar]
- 17.Court JA, Martin-Ruiz C, Graham A, Perry E. Nicotinic receptors in human brain: topography and pathology. J. Chem. Neuroanat. 2000;20:281–298. doi: 10.1016/S0891-0618(00)00110-1. [DOI] [PubMed] [Google Scholar]
- 18.Counts SE, et al. Alpha7 nicotinic receptor up-regulation in cholinergic basal forebrain neurons in Alzheimer disease. Arch. Neurol. 2007;64:1771–1776. doi: 10.1001/archneur.64.12.1771. [DOI] [PubMed] [Google Scholar]
- 19.Kalkman HO, Feuerbach D. Modulatory effects of alpha7 nAChRs on the immune system and its relevance for CNS disorders. Cell. Mol. Life Sci. 2016;73:2511–2530. doi: 10.1007/s00018-016-2175-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shytle RD, et al. Cholinergic modulation of microglial activation by alpha 7 nicotinic receptors. J. Neurochem. 2004;89:337–343. doi: 10.1046/j.1471-4159.2004.02347.x. [DOI] [PubMed] [Google Scholar]
- 21.Pavlov VA, Tracey KJ. The cholinergic anti-inflammatory pathway. Brain Behav. Immun. 2005;19:493–499. doi: 10.1016/j.bbi.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 22.Rosas-Ballina M, Tracey KJ. Cholinergic control of inflammation. J. Intern. Med. 2009;265:663–679. doi: 10.1111/j.1365-2796.2009.02098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagele RG, D’Andrea MR, Anderson WJ, Wang HY. Intracellular accumulation of beta-amyloid(1-42) in neurons is facilitated by the alpha 7 nicotinic acetylcholine receptor in Alzheimer’s disease. Neuroscience. 2002;110:199–211. doi: 10.1016/S0306-4522(01)00460-2. [DOI] [PubMed] [Google Scholar]
- 24.Gault J, et al. Genomic organization and partial duplication of the human alpha7 neuronal nicotinic acetylcholine receptor gene (CHRNA7) Genomics. 1998;52:173–185. doi: 10.1006/geno.1998.5363. [DOI] [PubMed] [Google Scholar]
- 25.Sinkus ML, et al. The human CHRNA7 and CHRFAM7A genes: a review of the genetics, regulation, and function. Neuropharmacology. 2015;96(Pt B):274–288. doi: 10.1016/j.neuropharm.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sinkus ML, et al. A 2-base pair deletion polymorphism in the partial duplication of the alpha7 nicotinic acetylcholine gene (CHRFAM7A) on chromosome 15q14 is associated with schizophrenia. Brain Res. 2009;1291:1–11. doi: 10.1016/j.brainres.2009.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lew AR, Kellermayer TR, Sule BP, Szigeti K. Copy number variations in adult-onset neuropsychiatric diseases. Curr. Genom. 2018;19:420–430. doi: 10.2174/1389202919666180330153842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szigeti K, et al. Ordered subset analysis of copy number variation association with age at onset of Alzheimer’s disease. J. Alzheimers Dis. 2014;41:1063–1071. doi: 10.3233/JAD-132693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swaminathan S, et al. Analysis of copy number variation in Alzheimer’s disease in a cohort of clinically characterized and neuropathologically verified individuals. PLoS ONE. 2012;7:e50640. doi: 10.1371/journal.pone.0050640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Lucas-Cerrillo AM, et al. Function of partially duplicated human alpha77 nicotinic receptor subunit CHRFAM7A gene: potential implications for the cholinergic anti-inflammatory response. J. Biol. Chem. 2011;286:594–606. doi: 10.1074/jbc.M110.180067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Araud T, et al. The chimeric gene CHRFAM7A, a partial duplication of the CHRNA7 gene, is a dominant negative regulator ofalpha7*nAChR function. Biochem. Pharmacol. 2011;82:904–914. doi: 10.1016/j.bcp.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsankov AM, et al. A qPCR ScoreCard quantifies the differentiation potential of human pluripotent stem cells. Nat. Biotechnol. 2015;33:1182–1192. doi: 10.1038/nbt.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fergus J, Quintanilla R, Lakshmipathy U. Characterizing pluripotent stem cells using the TaqMan(R) hPSC scorecard(TM) panel. Methods Mol. Biol. 2016;1307:25–37. doi: 10.1007/7651_2014_109. [DOI] [PubMed] [Google Scholar]
- 34.Liu Y, et al. Directed differentiation of forebrain GABA interneurons from human pluripotent stem cells. Nat. Protoc. 2013;8:1670–1679. doi: 10.1038/nprot.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y, et al. The duplicated α7 subunits assemble and form functional nicotinic receptors with the full-length α7. J. Biol. Chem. 2014;289:26451–26463. doi: 10.1074/jbc.M114.582858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Warren L, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma Y, et al. High-efficiency siRNA-based gene knockdown in human embryonic stem cells. RNA. 2010;16:2564–2569. doi: 10.1261/rna.2350710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang H, et al. Nicotinic acetylcholine receptor alpha7 subunit is an essential regulator of inflammation. Nature. 2003;421:384–388. doi: 10.1038/nature01339. [DOI] [PubMed] [Google Scholar]
- 39.Williams ME, et al. Ric-3 promotes functional expression of the nicotinic acetylcholine receptor alpha7 subunit in mammalian cells. J. Biol. Chem. 2005;280:1257–1263. doi: 10.1074/jbc.M410039200. [DOI] [PubMed] [Google Scholar]
- 40.Matta JA, et al. NACHO mediates nicotinic acetylcholine receptor function throughout the brain. Cell Rep. 2017;19:688–696. doi: 10.1016/j.celrep.2017.04.008. [DOI] [PubMed] [Google Scholar]
- 41.Nayak TK, Purohit PG, Auerbach A. The intrinsic energy of the gating isomerization of a neuromuscular acetylcholine receptor channel. J. Gen. Physiol. 2012;139:349–358. doi: 10.1085/jgp.201110752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Milescu LS, Nicolai C, Yildiz A, Selvin PR, Sachs F. Hidden Markov model applications in QuB: analysis of nanometer steps in single molecule fluorescence data and ensemble ion channel kinetics. Biophys. J. 2003;84:124a–124aa. doi: 10.1016/S0006-3495(03)74837-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu X, et al. Amyloid seeds formed by cellular uptake, concentration, and aggregation of the amyloid-beta peptide. Proc. Natl Acad. Sci. USA. 2009;106:20324–20329. doi: 10.1073/pnas.0911281106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gusev AG, Uteshev VV. Physiological concentrations of choline activate native alpha7-containing nicotinic acetylcholine receptors in the presence of PNU-120596 [1-(5-chloro-2,4-dimethoxyphenyl)-3-(5-methylisoxazol-3-yl)-urea] J. Pharmacol. Exp. Ther. 2010;332:588–598. doi: 10.1124/jpet.109.162099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pesti K, Szabo AK, Mike A, Vizi ES. Kinetic properties and open probability of alpha7 nicotinic acetylcholine receptors. Neuropharmacology. 2014;81:101–115. doi: 10.1016/j.neuropharm.2014.01.034. [DOI] [PubMed] [Google Scholar]
- 46.Dennis MY, Eichler EE. Human adaptation and evolution by segmental duplication. Curr. Opin. Genet. Dev. 2016;41:44–52. doi: 10.1016/j.gde.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kam TI, Gwon Y, Jung YK. Amyloid beta receptors responsible for neurotoxicity and cellular defects in Alzheimer’s disease. Cell. Mol. Life Sci. 2014;71:4803–4813. doi: 10.1007/s00018-014-1706-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.D’Andrea MR, Nagele RG. Targeting the alpha 7 nicotinic acetylcholine receptor to reduce amyloid accumulation in Alzheimer’s disease pyramidal neurons. Curr. Pharm. Des. 2006;12:677–684. doi: 10.2174/138161206775474224. [DOI] [PubMed] [Google Scholar]
- 49.Wang HY, Li W, Benedetti NJ, Lee DH. Alpha 7 nicotinic acetylcholine receptors mediate beta-amyloid peptide-induced tau protein phosphorylation. J. Biol. Chem. 2003;278:31547–31553. doi: 10.1074/jbc.M212532200. [DOI] [PubMed] [Google Scholar]
- 50.Baranello RJ, et al. Amyloid-beta protein clearance and degradation (ABCD) pathways and their role in Alzheimer’s disease. Curr. Alzheimer Res. 2015;12:32–46. doi: 10.2174/1567205012666141218140953. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.