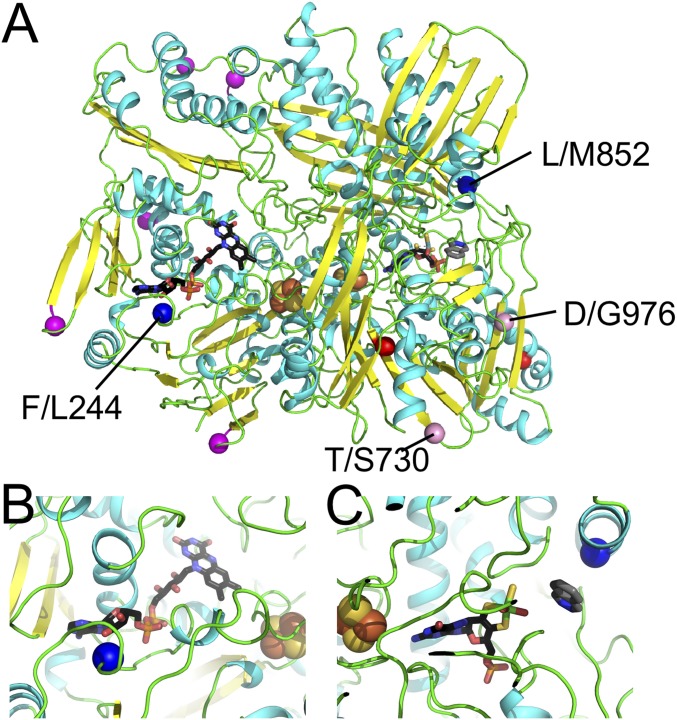
Fig. 5.
An aldehyde oxidase shows elevated divergence between EGM and AGM populations. (A) L. dispar aldehyde oxidase (ldi971.1) modeled on a human aldehyde oxidase template (4UHW) is depicted. Colors indicate secondary structure: helix (cyan), strand (yellow), and loop (green) with FeS centers (orange/yellow spheres), FAD redox cofactor (black stick), molybdenum cofactor (black stick), and substrate (gray stick). Population-specific sites map to the model surface (G/S17, E/K304, C/Y366, L/Q511, C/F516, T/S730, and D/G976; Cα positions are shown with magenta/pink spheres), with two of the surface residues contributing to the dimer interface (D/G976 and T/S730; Cα positions shown with pink spheres). Two sites form the hydrophobic core of their respective subdomains (T/A725 and V/I1056; Cα positions shown with red spheres). (B and C) Zoom-in of two polymorphic residues that map near active sites (blue spheres): F/L244, which lines the FAD-binding site (black stick) (B), and L/M852, which lines the substrate-binding site (gray stick) in the molybdenum cofactor (black stick)-binding domain (C).