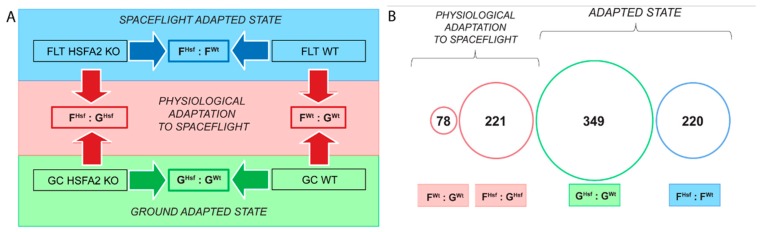
Figure 2.
Graphical presentation of the dimensions used in the microarray data analysis and the results of thereof. (A) HSFA2 KO and WT mark the gene expression profiles for respective cell samples. Solid arrows represent the direction of comparison of the gene expression profiles. Red box and arrows indicate the first dimension of data analysis—gene expression profiles of spaceflight-adapted state to ground-adapted state for each of the two cell lines, thereby characterizing the physiological adaptation of each genotype to spaceflight. Green box and arrow indicate part of the second dimension of data analysis—a comparison of gene expression profiles between the two genotypes in the ground-adapted state, thereby defining the genes uniquely required by the two genotypes for ground-adapted state. Blue box and arrow indicate part of the second dimension of data analysis—a comparison of gene expression profiles between the two genotypes during spaceflight-adapted state, thereby defining the genes uniquely required by the two genotypes for spaceflight-adapted state; (B) A proportional graphical presentation of the significantly differentially expressed genes identified in each comparison group: 78 genes of the physiological adaptation to spaceflight in WT cells obtained from FWt : GWT; 221 genes of the physiological adaptation to spaceflight in HSFA2 KO cells obtained from FHsf : GHsf; 349 genes of the ground-adapted state between HSFA2 KO and WT cells obtained from GHsf : GWt group; 220 genes of the spaceflight-adapted state between HSFA2 KO and WT cells obtained from FHsf : FWt.