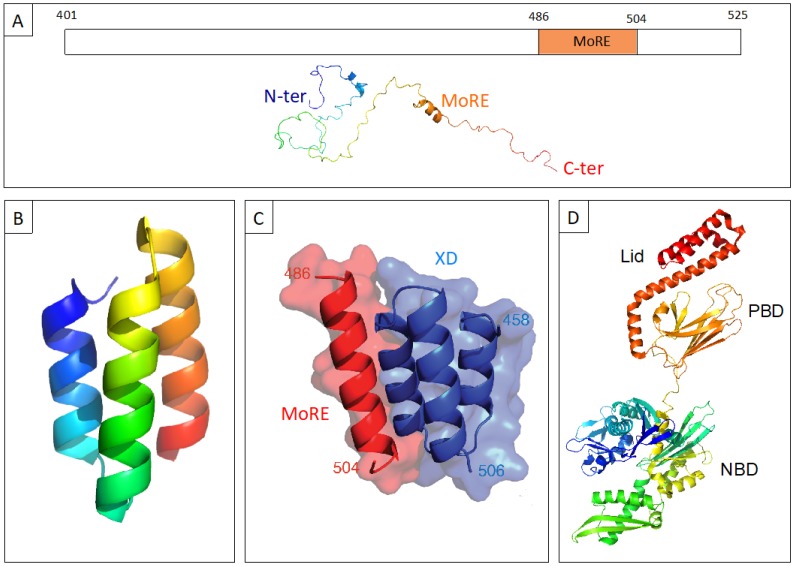
Figure 1.
(A) Schematic representation of the C-terminal domain of the nucleoprotein (N) of measles virus (MeV) NTAIL (upper panel) and cartoon representation of an NTAIL conformer generated using Flexible-Mecano [49]. (B) Ribbon representation of the crystal structure of the C-terminal X domain of the phosphoprotein of MeV XD (PDB code 1OKS). (C) The structure of the chimeric construct made of MeV XD (blue) and of the molecular recognition element MoRE of NTAIL (red) (PDB code 1T6O). (D) Cartoon representation of the crystal structure of hsp70 based on PDB codes 1HJO and 4JNF. The relative orientation of the two hsp70 domains (i.e., amino acids 3 to 382 and amino acids 389 to 610) is based on the structure of a form encompassing residues 1 to 554 (PDB code 1YUW). The three constituent domains of hsp70, i.e., nucleotide binding domain (NBD, aa 1 to 384), peptide binding domain (PBD, aa 384 to 543) and “lid” (aa 543 to 641) (see [18] and references therein cited) are highlighted.