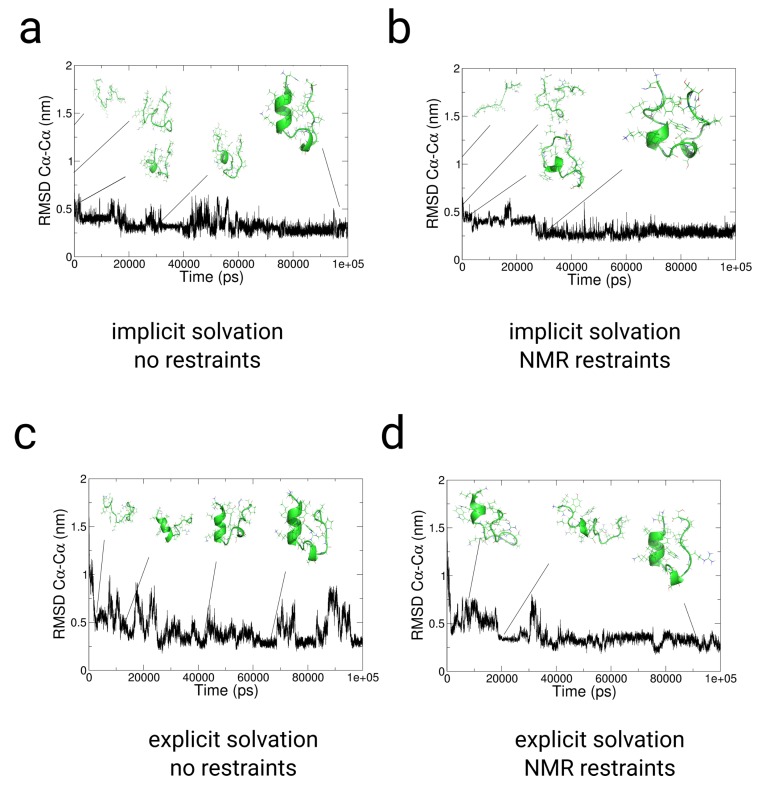
Figure 3.
Results from path-sampling simulations with and without restraints from NMR-NOESY measurements of TrpCage starting from an extended conformation [93] in implicit (a,b) and explicit solvent (c,d). Panels (a,c) Results from un-restrained folding simulations of TrpCage in implicit and explicit solvent. Panels (b,d) Results from folding simulations of TrpCage with NMR restraints in implicit and explicit solvent. (a–d) to the backbone of NMR-model #1 of PDB: 1l2y. In each of the cases, we observe folding to lower than 0.23 nm. The application of the set of restraints (b,d) changes the partition of conformations and prevents spontaneous unfolding to configurations at greater than 0.4 nm as we observe in path-sampling simulations without restraints (a,c).