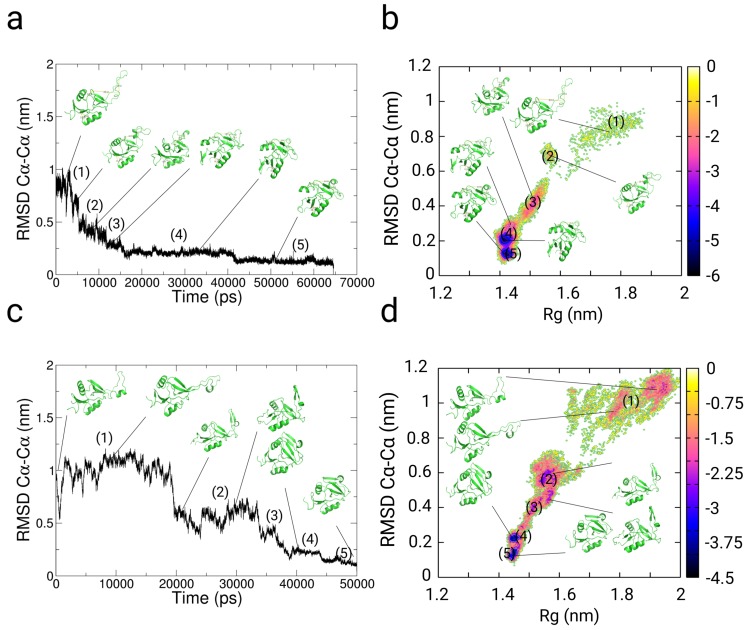
Figure 5.
Results from restrained simulations of the Killer Cell Lectin-like Receptor Subfamily B Member 1A (NKR-P1A) along multiple path-dependent biasing variables with chemical cross-linking restraints from ref. [89] (PDB: 3m9z as starting structure). (a,b) Results from enhanced restrained simulations using the path-dependent biasing approach in implicit solvent. (c,d) Results from enhanced restrained simulations along multiple path-dependent biases in explicit solvent. (a,c) to the final structure of each simulation. (b,d) Free energy landscape as function of and the radius of gyration (Rg). Energy values on the color bar are in units of . The protein relaxes quickly out of the extended conformation in both simulations within few ns and adopts 5 different compact states, where the loop region (residues: Arg157-Ser188) are packed to the -strand interface (residues: Ser188-Asn203) and parts of the second N-terminal helix (residues: Gln135-Ile145) of the monomer.