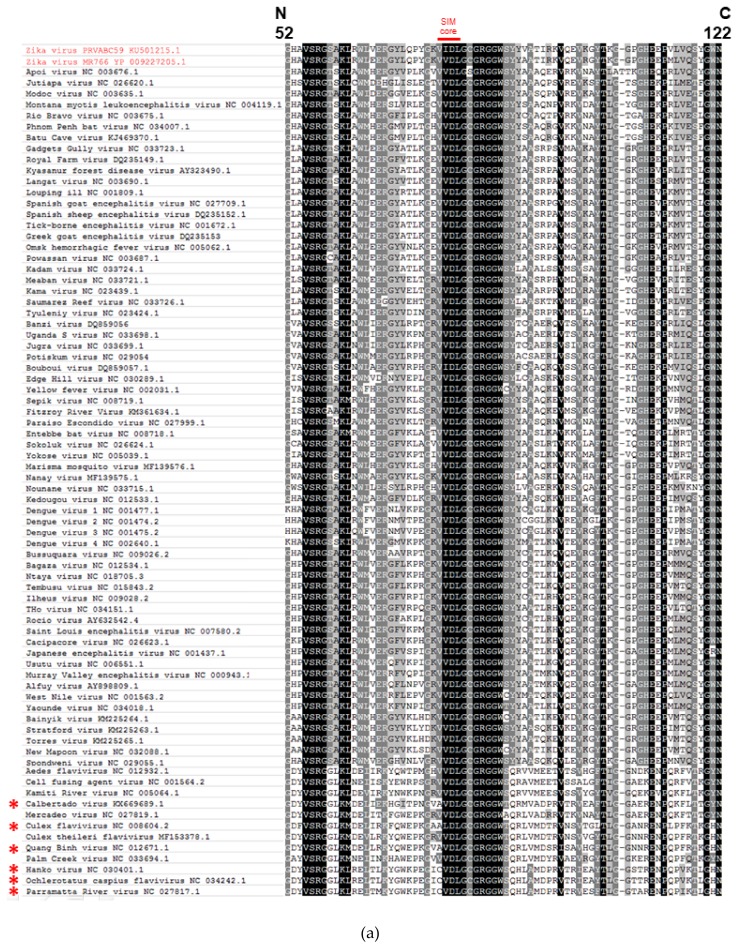
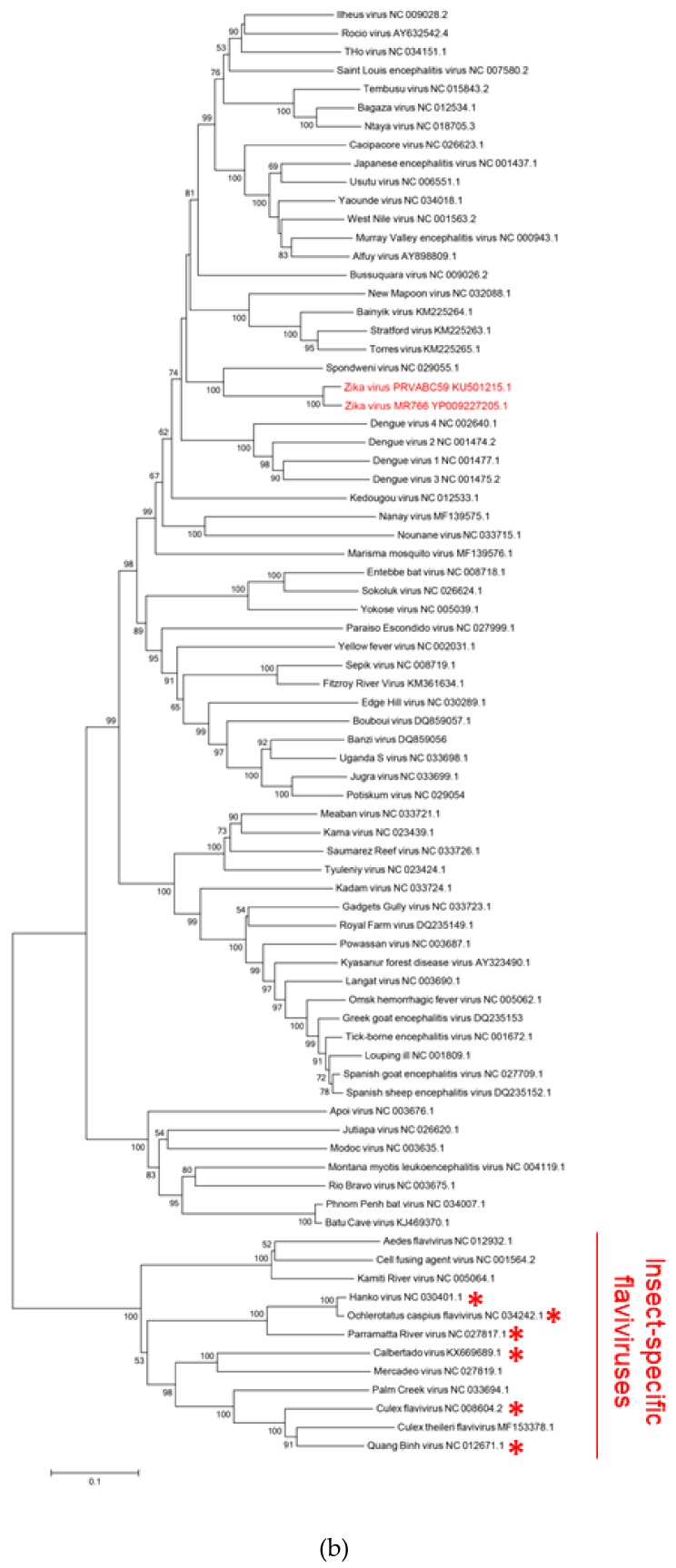
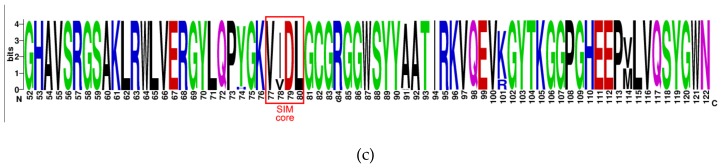
Figure 1.
The putative SUMO-interacting motif (SIM) at the N-terminal domain of non-structural 5 (NS5) protein is highly conserved among flaviviruses and among pre-epidemic and epidemic Zika virus (ZIKV) strains. (a) Multiple sequence alignment of the NS5 amino acid sequences around the putative SIM of 78 representative flaviviruses with complete genome sequences available in GenBank (accessed on 1 May 2018) showing conserved amino acid sequences (VIDL or VVDL) of the putative SIM core among most (72/78, 92.3%) of the flaviviruses (labelled as “SIM core” in red). The amino acid positions (52 to 122) represent those of the epidemic ZIKV-PR (Puerto Rico strain PRVABC59, accession number KU501215). (b) Phylogenetic analysis of the NS5 protein of the 78 flaviviruses. The tree was constructed by the neighbor-joining method using MEGA 6.0 software, with bootstrap values being calculated from 500 trees. Bootstrap values lower than 50 were hidden. All flavivirus strains are labeled with their names followed by their accession numbers. In both (a) and (b), the epidemic ZIKV-PR and pre-epidemic ZIKV-MR766 (accession number YP_009227205) are highlighted in red. The 6 insect-specific flaviviruses which have different putative SIM core amino acids of CVDL, AVDL, or ALDL are marked by red asterisks. (c) The Sequence logo of the partial NS5 protein amino acid sequences (amino acid positions 52 to 122) of 414 ZIKV strains with complete genomes available in GenBank (accessed on 9 January 2018) show that the putative SIM at the N-terminal domain of the NS5 protein is highly conserved among pre-epidemic and epidemic ZIKV strains. The red box represents the putative SIM core at amino acid positions 77 to 80. The height of each stack is measured in “bits” of information, with each amino acid ordered from the most frequent to least frequent.