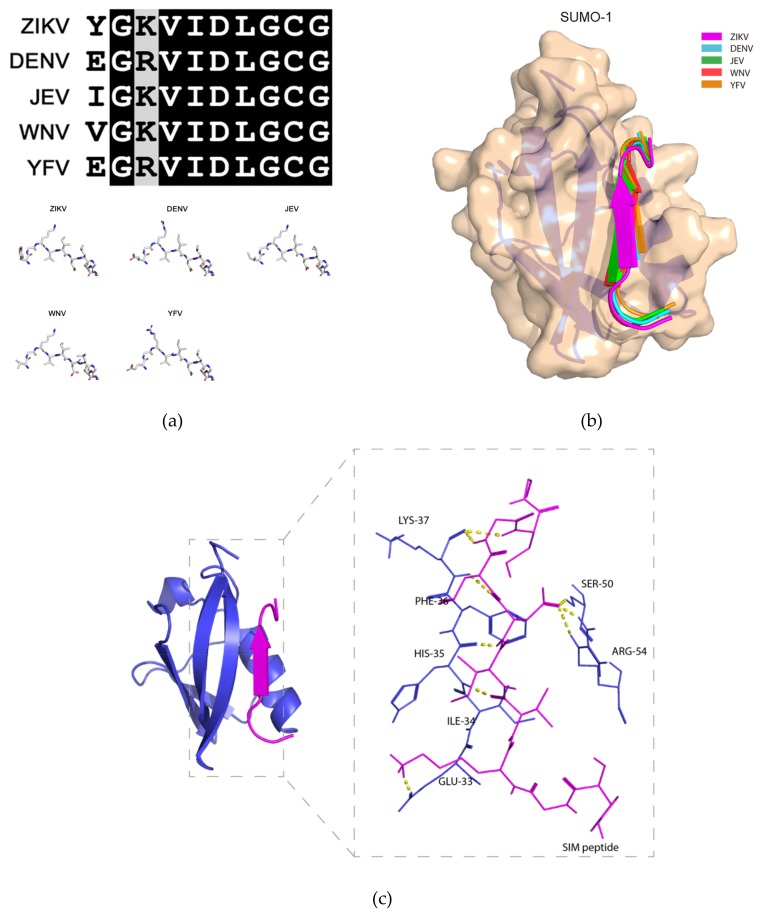
Figure 2.
Molecular docking model of the binding between the putative non-structural 5 (NS5) SUMO-interacting motifs of flaviviruses and the SUMO-1 protein. (a) Top panel: multiple sequence alignment of the amino acid sequences of the putative NS5 protein SUMO-interacting motifs (SIM) of Zika virus (ZIKV), dengue virus (DENV) (serotype 3), Japanese encephalitis virus (JEV), West Nile virus (WNV), and yellow fever virus (YFV). Bottom panel: Stick representation of the structural similarities among the 5 flaviviruses’ putative NS5 SIM peptides. (b) Schematic representation of the binding between the SUMO-1 protein and the 5 flaviviruses’ putative NS5 SIM peptides. The SUMO-1 protein is shown in wheat and the NS5 SIM peptides are shown in different colors. (c) Ribbon representation showing the interacting amino acid residues of the putative ZIKV NS5 SIM peptide and the active sites of the SUMO-1 protein. The putative ZIKV NS5 SIM peptide and SUMO-1 protein are displayed in magenta and blue, respectively. The interacting residues are shown as sticks with hydrogen bonds represented by yellow dashed lines.