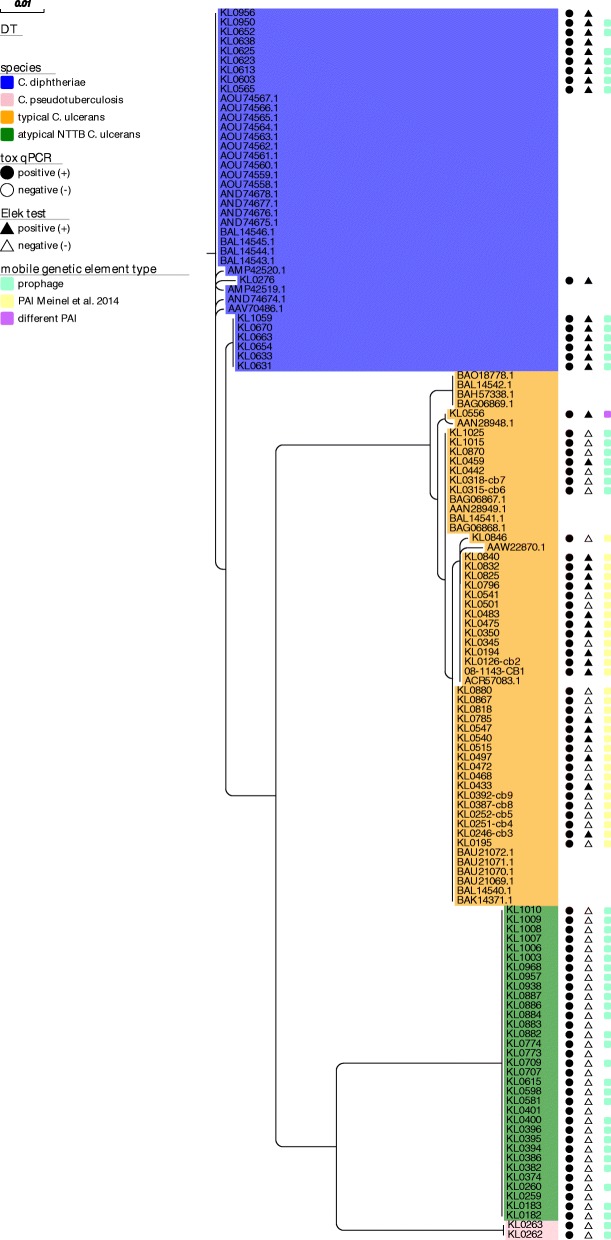
Fig. 1.
Maximum likelihood phylogeny of translated DT sequences from different Corynebacterium species. Translated DT sequences of 90 of 91 toxigenic Corynebacterium spp. isolates were extracted, aligned together with 39 NCBI derived DT sequenced from C. diphtheriae and C. ulcerans and a maximum likelihood tree was built. Species affiliation is indicated by background coloring: C. diphtheriae = blue, C. pseudotuberculosis = pink, typical C. ulcerans = orange, atypical NTTB C. ulcerans = green. Results of tox qPCR and Elek tests are indicated by circle and triangle shaped bars, respectively: positive = black, negative = white. The type of detected tox-surrounding genetic mobile element is indicated by colored boxes on the right side of the tree: prophage = turquoise, alternative PAI described in [7] = yellow, different PAI than the one described in [7] = purple