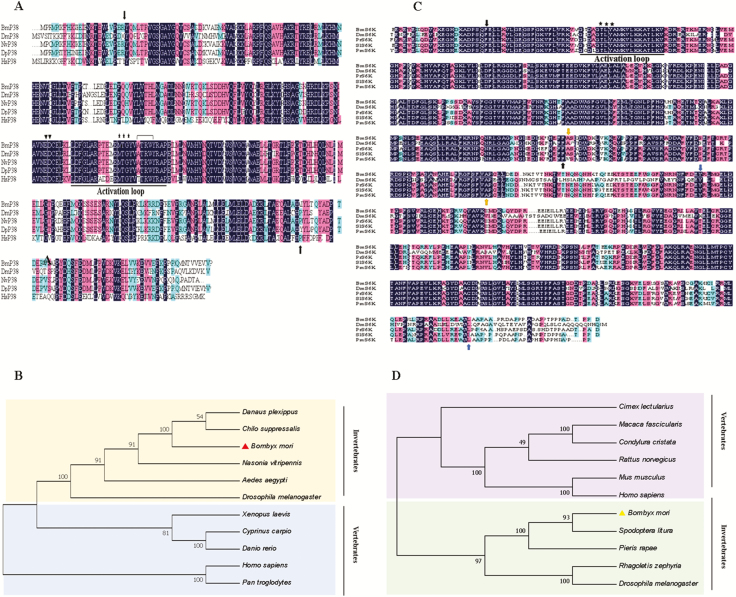
Fig. 3.
(A) Multiple sequences alignment of Bmp38 with other known organisms p38 homolog. A hundred percent conserved amino acids are shaded in black. The domain is represented by black arrows. The activation loop is underlined, ED site is shown by inverted triangles, TGY motif is shown by stars, and substrate site is represented by inverted bracket. Organisms used for multiple sequence alignment are Bombyx mori (NP_001036996.1), Drosophila melanogaster (AAB97138.1), Nasonia vitripennis (XP_001136337.1), Danaus plexipus plexipus (OWR52323.1), and Homo sapiens (AAC51758.1). (B) Multiple sequences alignment of BmS6K with other known organisms S6K homolog. The positions of three different domains are shown by different colors arrows (black, yellow, and blue). The activation loop is underlined, TYL motif is shown by stars. Bombyx mori (NP_001189457.1), D. melanogaster (AAA50509.1), Pieris rapae (XP_022122087), Spodoptera litura (XP_022827882.1), and Papilio machaon (KPJ06800.1) were used for alignment. (C) Phylogenetic tree of Bmp38 with other organisms. Bombyx mori p38 is shown with red color triangle. Organisms used to construct p38 phylogenetic tree are B. mori (NP_001036996.1), D. melanogaster (AAB97138.1), Aedes aegypti (XP_001653239.1), N. vitripennis (XP_001136337.1), D. p. plexipus (OWR52323.1), Rattus norvegicus (AAB51285.1), H. sapiens (AAC51758.1), Pan troglodytes (NP_00102926.1), Chilo suppressalis (AN046359.1), Xenopus laevis (NP_001165343.1), Cyprinus carpio (BAA96415.1), and Danio rerio (NP_571797.1). (D) Phylogenetic tree of BmS6K with other organisms. Bombyx mori S6K is shown with yellow color triangle. Organisms used to construct phylogenetic trees are B. mori (NP_001189457.1), D. melanogaster (AAA50509.1), R. norvegicus (NP_112369.1), Cimex lectularis (XP_024085928.1), Macaca fascicularis (XP_005593208.1), Condylura cristata (XP_004689960.1), Mus musculus (NP_035429.1), H. sapiens (BAD92353.1), S. litura (XP_022827882.1), Pieris rapae (XP_022122087), and Rhagoletis zephyria (XP_ 017483318.1).