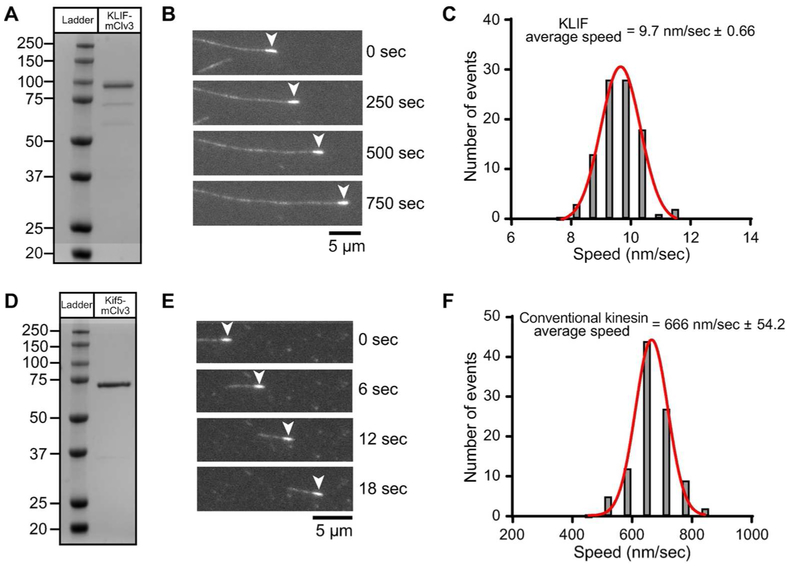
Figure 8.
In vitro characterization of expressed KLIF motor domain. A,D) Lane 1, molecular weight protein markers on a 4-12% SDS PAGE gradient gel stained with Coomassie. Lane 2, bacterial expressed motor domain of KLIF (a.a. 1-672) or conventional kinesin Kif5 (a.a. 1-407) fused at the C-terminus with the GFP variant, mClover3 and His8 tag, purified by nickel affinity and size exclusion chromatography. B,E) Epifluorescence images showing movement of polarity marked microtubules on KLIF-mClv3 or Kif5-mClv3 using an in vitro motility assay. The minus-end of the microtubule (arrowhead) is marked with a heavily rhodamine-labeled microtubule seed. Movement of microtubules with the minus-end leading indicates that KLIF and Kif5 are plus-end directed motors. Kif5 has a well-established role in plus-end directed cargo transport. C) Representative speed histogram showing that KLIF moves microtubules with an average velocity of 9.7 ± 0.66 nm/sec (N = 100) at 1 mM MgATP concentrations. Data is representative of 3 independent experiments and two protein preparations. F) Representative speed histogram showing that conventional kinesin moves microtubules with an average velocity of 666 ± 54.2 nm/sec (N = 100) at 1 mM MgATP concentrations. Data is representative of 2 independent experiments and one protein preparation.