Abstract
Androgen Receptor (AR) transcriptional activity contributes to prostate cancer (PCa) development and castration resistance. The growth and survival pathways driven by AR remain incompletely defined. Here we found PDCD4 to be a new target of AR signaling, and a potent regulator of PCa cell growth, survival, and castration resistance. The 3’ untranslated region of PDCD4 is directly targeted by the androgen-induced miRNA, miR-21. Androgen treatment suppressed PDCD4 expression in a dose-responsive and miR-21-dependent manner. Correspondingly, AR inhibition dose-responsively induced PDCD4 expression. Using data from PCa tissue samples in the Cancer Genome Atlas (TCGA), we found a significant and inverse correlation between miR-21 and PDCD4 mRNA and protein levels. Higher Gleason grade tumors exhibited significantly higher levels of miR-21, and significantly lower levels of PDCD4 mRNA and protein. PDCD4 knockdown enhanced androgen-dependent cell proliferation and cell cycle progression, inhibited apoptosis, and was sufficient to drive androgen-independent growth. On the other hand, PDCD4 over-expression inhibited miR-21 mediated growth and androgen-independence. The stable knockdown of PDCD4 in androgen-dependent PCa cells enhanced subcutaneous tumor take rate in vivo, accelerated tumor growth, and was sufficient for castration resistant tumor growth.
Keywords: Prostate Cancer, Androgen Receptor, microRNA, Castration Resistance
INTRODUCTION
Prostate cancer (PCa) is the most frequent urological malignancy and a leading cause of cancer death in men worldwide (1). There are over 160,000 estimated new cases of PCa in the United States in 2018, with more than 29,000 predicted deaths (2). Androgen deprivation therapy (ADT) is widely utilized as a standard treatment for locally advanced and metastatic PCa (3). Although many patients initially respond to ADT, they almost invariably relapse and develop castration resistant prostate cancer (CRPC), often through the re-activation of androgen receptor (AR) signaling (4,5). CRPC eventually causes cancer death, even after life-prolonging therapies such as cabazitaxel, sipuleucel-T, abiraterone, enzalutamide, and Ra-223 (6). Thus, there is a need to identify mechanisms of AR-induced tumor growth and castration resistance, and to establish new therapeutic targets and strategies for the treatment of CRPC.
MicroRNAs (miRNA) are a class of non-coding RNA that post-transcriptionally suppress gene expression through seed-pairing interactions with specific messenger RNAs (mRNAs) (7). MicroRNA-21 (hsa-miR-21-5p, miR-21) is a well-recognized and cancer-associated miRNA with proven oncogenic activity (8). Levels of miR-21 are frequently elevated in most carcinomas, when compared to non-malignant tissue, including in PCa and CRPC (9–11). Several cancer-related pathways, including AP-1, RAS, IL-6, and TGF-β, induce the expression of miR-21 (12–15). We previously discovered that the activated AR also directly induces miR-21 transcription, and that elevated miR-21 expression enhances androgen-dependent PCa growth and is sufficient to drive castration resistant PCa cell growth (16). However, the mechanisms of miR-21 mediated castration resistance remain undefined.
In the current study, we examined several miR-21 target genes as potential regulators of PCa growth. We found one target, PDCD4 (17,18), to be a potent suppressor of PCa growth and survival. Our results show that androgen signaling suppresses PDCD4 expression, through miR-21, and that PDCD4 knockdown enhances cell proliferation, reduces apoptosis, augments tumorigenesis, and drives castration resistance. We also found that localized, high Gleason grade PCa exhibits low levels of PDCD4 expression, and high levels of miR-21 expression. These results implicate PDCD4 as a potential regulator of PCa aggressiveness and therapeutic resistance.
MATERIALS AND METHODS
Reagents and chemicals
Methyltrienolone (R1881) (Perkin-Elmer, Waltham, MA, USA) was dissolved in ethanol and Bicalutamide (Sigma-Aldrich, St Louis, MO, USA) was dissolved in dimethyl sulfoxide and stored at −20 °C. pCMV6-XL5-PDCD4 (SC111794) was purchased from OriGene Technologies Inc. (Rockville, MD, USA). pCMV6-EV was generated by PDCD4 removal and self-ligation. FlexiTube siRNA were purchased from Qiagen (Valencia, CA, USA). Details of siRNAs, shRNAs, plasmids, and miRNAs are provided in Supplementary Table S1.
Cell culture
LNCaP and HEK 293 cells were purchased from ATCC (Manassas, VA, USA). LAPC4 cells were provided by Dr. John Isaacs (Johns Hopkins, Baltimore, MD). All cell lines were authenticated using the Power Plex 16 HS System (Promega Corporation, Madison, WI, USA) and routinely verified to be mycoplasma-free. Cell cultures were maintained at 37 °C in 5 % CO2.
Reporter plasmids
The 3’UTR regions of miR-21 targets were amplified from HEK 293 genomic DNA (human embryonic kidney cells) by PCR. Primer sequences are provided in Supplementary Table S2. PCR products were inserted into either the SpeI and HindIII or SacI and PmeI cloning sites of the pMIR-REPORT luciferase expression reporter vector (Ambion, Austin, TX, USA) to generate pMIR-UTR and mutant pMIR-UTR. The miR-21 Reporter control consists of four perfect miR-21 binding sites cloned into the 3’UTR region of the pMIR-report vector.
Dual Luciferase Reporter Assays
HEK 293 cells grown in 96-well plates were transfected with 25 nM hsa-miR-21 mimic or negative control mimic, 100 ng luciferase-UTR reporter plasmid, 10 ng of reference pRL-CMV Renilla reporter plasmid and OPTI-MEM to give a final volume of 100 μl. Luciferase activity was assayed 24-48 hours post-transfection using the dual luciferase assay and a Wallac Microbeta Luminometer. Firefly luciferase activity was normalized by Renilla luciferase. In each case, quadruplicate experiments were performed in parallel and repeated at least three times and up to nine times.
Transfection and androgen treatment or inhibition
Transient transfections applied 20 nM of miRNAs, miRNA inhibitors, or siRNAs, or plasmid vector (100 ng for 96-well, 1000 ng for 6-well), using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). To modulate androgen signaling, cells were grown or treated in complete medium (CM), consisting of RPMI1640 with 10 % fetal bovine serum, or androgen depleted medium (ADM), consisting of phenol red-free RPMI 1640 media supplemented with 10 % charcoal-stripped serum (GE Healthcare Life Sciences, Logan, UT, USA). AR activation was achieved with R1881 and AR inhibition was achieved with Bicalutamide (Sigma-Aldrich).
Cell proliferation analysis
Transiently transfected or stable cells were plated 48 h after transfection into different media conditions (CM, ADM, 10 μM of Bicalutamide). Cell viability was evaluated by MTS assay (Promega) 7 days after plating, or by MLuc Cell Viability Assay, as previously described (19).
Gene expression analysis
Clinical parameters, Illuminahiseq_mirnaseq-miR_gene_expression, Mda_rppa_core-protein_normalization, and Illuminahiseq_rnaseqv2-RSEM_genes_normalized data were downloaded from the TCGA data version 2016_01_28 for PRAD through The Broad Institute TCGA GDAC Firehose (20). The correlation between the normalized expression of PDCD4 mRNA, PDCD4 protein, and hsa-miR-21 were determined by Patient Sample ID, and association with radical prostatectomy Gleason Score by Patient ID.
Western blot analysis
Cells were lysed in RIPA (Sigma-Aldrich) lysis buffer and extracts separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred onto nitrocellulose membranes. Membranes were blocked with odyssey blocking buffer overnight at 4 °C with antibodies against PDCD4 (1:1000, D29C6, Cell Signaling Technology, Danvers, MA, USA), c-Myc (1:1000, Y69, Abcam, Cambridge, MA, USA), Cyclin E1 (1:1000, HE12, Cell Signaling Technology), ACTB (1:10000, AC-15, Sigma-Aldrich) and GAPDH (1:10000, G9545, Sigma-Aldrich). Proteins were detected by IRDye secondary antibody (LI-COR, Lincoln, NE) at room temperature for approximately 1 h. Images were analyzed with the Odyssey infrared imaging system (LI-COR). Signals for each protein were normalized to ACTB or GAPDH.
Reverse transcription and qPCR
Total RNA was isolated with Trizol reagent (Invitrogen). cDNAs were synthesized with 1 μg RNA using the QuantiTect Reverse Transcription Kit (Qiagen, Valencia, CA, USA). qPCR applied 1 μl diluted cDNA (1:10), The SYBR GreenER™ qPCR SuperMix for ABI PRISM™ Instrument (Invitrogen), ABI Prism 7900HT (Applied Biosystems, Forster City, CA, USA). The primer sequences of each gene are shown in Supplementary Table S2.
Generation of stable cell lines
To generate stable PDCD4 knockdown cell lines, cells were transduced with lentiviral supernatants of pLKO.1-non-target shRNA control or pLKO.1-shPDCD4 (Supplementary Table S1) and selected by 2 μg/ml puromycin.
Flow cytometry
For cell cycle analysis, cells were incubated in CM or ADM for 72 h and trypsinized. Cells were fixed with 70 % ethanol overnight at 4°C. Fixed cells were washed twice with cold phosphate-buffered saline (PBS) and incubated with 100 μg/mL RNase A and 50 μg/mL propidium iodide (PI) at 37 °C for 1 h. Samples were analyzed on the S3e cell sorter (Bio-Rad) and cell cycle distribution was determined using the FlowJo software. Detection of apoptotic cells was performed using the Annexin V Apoptosis Detection Kit (Invitrogen). Cells were treated with different media conditions (CM, ADM) for 72 h, and harvested and washed with cold PBS, incubated with Annexin V-FITC and propidium iodide in the dark at room temperature for 15 min and analyzed by flow cytometry.
Tumor xenografts
Animal studies were performed according to the protocols approved by the Animal Care and Use Committee at Johns Hopkins University. 6-week-old male athymic nu/nu mice (Charles River Laboratories Inc., Frederick, MD, USA) were inoculated subcutaneously in the right dorsal flank with 5 × 106 cells. Cells were suspended in equal volumes with Matrigel (Corning, Corning, NY, USA). Surgical castration was performed through bilateral orchiectomy at average tumor volume of 350 mm3 for LNCaP or 380 mm3 for LAPC4. Tumors were measured every other day and tumor volume (mm3) was calculated by length × width × height × 0.52.
Statistical analysis
The results are reported as the mean ± standard deviation (S.D.) or standard error (S.E.). Statistical analyses applied GraphPad Prism (La Jolla, CA, USA). The differences between groups were evaluated by two-tailed, unpaired Student’s t-test. Tumor volume over time was evaluated by two-way ANOVA with Bonferroni post-test for each time point. P < 0.05 was considered statistically significant. Correlative expression was determined by Spearman’s correlation analysis and association with Gleason Score by One-way ANOVA and Bonferroni’s Multiple Comparison Test.
RESULTS
Identification of miR-21 target genes that mediate androgen independence
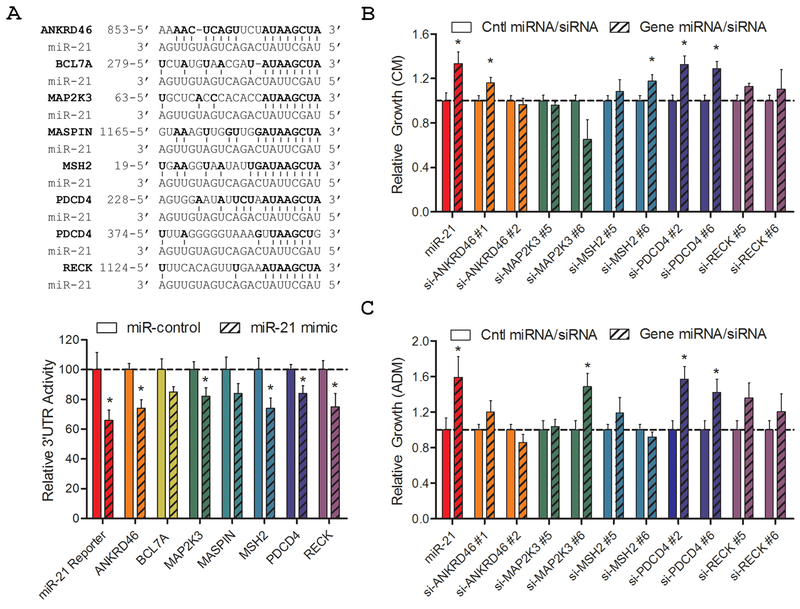
We examined seven previously identified miR-21 targets (ANKRD46, BCL7A, MAP2K3, MASPIN, MSH2, PDCD4, and RECK) as potential regulators of PCa growth and castration resistance (16,18,21–26). The predicted miR-21 binding regions for each gene transcript were subcloned downstream of the firefly luciferase gene to test for miR-21 regulation in 3’UTR reporter assays (Fig. 1A, Top). A positive control 3’UTR reporter construct, miR-21 Reporter, was engineered to contain four consecutive miR-21 binding sites. Each 3’UTR reporter was co-transfected into HEK 293 cells with miR-21 mimic, or control miRNA, and luciferase activity measured after two days. Six of the eight 3’UTR reporters were significantly suppressed by miR-21, verifying these genes as direct miR-21 targets (Fig. 1A, Bottom).
Figure 1. Screening for miR-21 target genes that promote prostate cancer growth and androgen independence.
A, 3’UTR reporter analysis of reported miR-21 target genes. Top: Alignment of reported miR-21 target genes and miR-21 binding sites. Numbers indicate nucleotide site within mRNA. Bottom: HEK 293 cells were transfected with the 3’UTR report vector for each gene, miR-21 or control mimic, and reference Renilla luciferase. Luciferase activity is normalized to Renilla and relative to miR-control. Bars represent mean + S.E. of at least five independent quadruplicate experiments. B, Androgen-dependent cell growth in CM as measured by MLuc Viability Assay, seven days after miR-21 or siRNA transfection. C, Androgen-independent cell growth in ADM as measured by MLuc Viability Assay, seven days after miR-21 or siRNA transfection. Growth is relative to control miRNA or control siRNA for each gene. Bars represent mean + S.E. of two independent triplicate experiments. *, p < 0.05.
Two separate siRNAs were then selected for each verified miR-21 target gene. Each siRNA was separately transiently transfected into the androgen-dependent cell line, LNCaP-MLuc. This cell line expresses secreted Metridia luciferase under the control of the β-actin promoter and enhancer, allowing non-invasive quantification of viable cell number over time by measuring relative MLuc activity in the conditioned media (19). We measured the androgen-dependent growth of siRNA-transfected cells after seven days in complete media (CM), which includes androgens from serum. Only one miR-21 target gene, PDCD4, showed enhanced androgen-dependent growth in CM when targeted by two different siRNAs (Fig. 1B). We also measured the effects of each siRNA on androgen-independent cell growth, in androgen-depleted media (ADM), where androgens were charcoal-stripped from serum and phenol red was excluded from the media. Again, only one miR-21 target gene, PDCD4, drove androgen-independent growth when targeted by two different siRNAs (Fig. 1C). These results implicate PDCD4 as a possible regulator of PCa growth and castration resistance.
PDCD4 expression is suppressed by miR-21 and Androgen Receptor Signaling
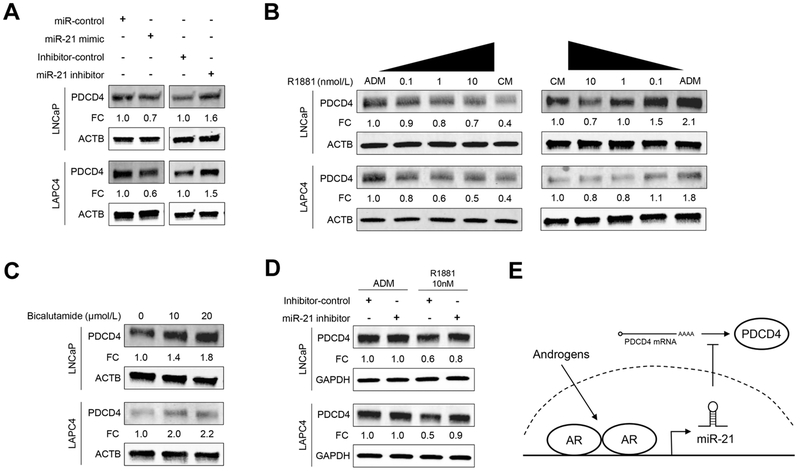
We then sought to study endogenous PDCD4 protein regulation in two established androgen-sensitive cell lines: LNCaP and LAPC4. Transient transfection with miR-21 mimics reduced PDCD4 protein expression in both cell lines by 30-40% (Fig. 2A). We then inhibited endogenous miR-21 by transfecting miR-21 inhibitors, which increased PDCD4 protein expression by 50-60%. PDCD4 expression was similarly suppressed by miR-21 mimics, and induced by miR-21 inhibitors, in normal prostate epithelial cells (Supplementary Fig S1). Understanding that the AR directly activates the miR-21 promoter (16,27), we hypothesized that androgen treatment would suppress PDCD4 protein expression. To test this, cells were grown in ADM for 24 h and then treated with increasing levels of androgen (0.1-10 nM R1881). As hypothesized, androgen treatment caused a dose-responsive decrease in PDCD4 protein expression in both LNCaP and LAPC4 cells (Fig. 2B, left panel). In a complementary experiment, cells were grown in CM for 24 h, and then treated with media containing decreasing levels of androgens (10-0.1 nM R1881), or ADM. Androgen deprivation produced a dose-responsive increase in PDCD4 protein expression (Fig. 2B, right panel). Consistent with this, PDCD4 mRNA expression was elevated upon androgen deprivation, and PDCD4 mRNA expression was reduced by miR-21 mimic transfection (Supplementary Fig. S2).
Figure 2. Androgen signaling suppresses PDCD4 expression through miR-21.
A, LNCaP and LAPC4 cells were transfected with 20 nM of miR-21 mimic or inhibitor for 48 h and PDCD4 expression quantified by WB, normalized to ACTB. B, Cells were grown in ADM for 24 h and then stimulated with indicated amounts of androgen (R1881) or CM for 24 additional hours (left panel); or cells were grown in CM condition for 24 h and then incubated with indicated concentration of R1881 or ADM for 24 h (right panel). PDCD4 expression quantified by WB, normalized to ACTB. C, Cells were incubated in CM for 24 h, then treated for 24 h with Bicalutamide or vehicle control. PDCD4 expression quantified by WB, normalized to ACTB. D, Cells were transfected with 20 nM of miR-21 inhibitor or control. The next day cells were exposed to ADM, or ADM supplemented with 10 nM R1881, for 24 h. PDCD4 expression quantified by WB, normalized to GAPDH. FC, Fold Change relative to control. E, Schematic of suppressed PDCD4 expression through androgen signaling and miR-21.
To further study PDCD4 suppression by AR signaling, we treated cells with the small molecule AR inhibitor, Bicalutamide. Bicalutamide treatment caused a dose-responsive increase in PDCD4 protein expression in both LNCaP and LAPC4 cell lines (Fig. 2C). Finally, to determine if androgens suppressed PDCD4 through endogenous miR-21, we transfected androgen-deprived cells with miR-21 inhibitors or control miRNA inhibitors, and then treated cells with androgen (10 nM R1881). Consistent with the above results, androgen treatment suppressed PDCD4 protein expression in control-transfected cells by 40-50 % (Fig. 2D, ADM Inhibitor-control vs R1881 Inhibitor-control). However, when miR-21 was blocked, androgens only suppressed PDCD4 expression by 10-20% (Fig. 2D, R1881 Inhibitor-control vs R1881 miR-21-inhibitor). The miR-21 inhibitor did not affect PDCD4 expression in the absence of androgens. Collectively, these results indicate that androgens suppress PDCD4 expression in PCa cells through miR-21 (Fig. 2E).
miR-21 and PDCD4 expression are inversely correlated in human prostate cancer and altered expression is associated with high tumor Gleason Score
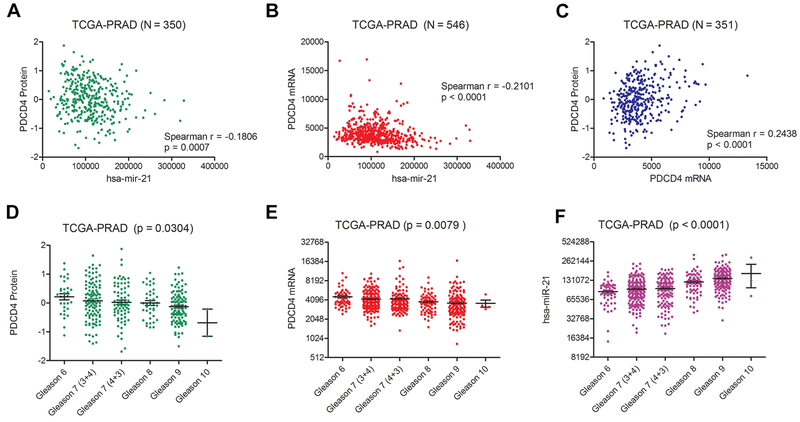
To determine the relationship between miR-21 and PDCD4 expression in human PCa tissues, we performed Spearman correlation analyses using Level 3 miRNA, mRNA, and protein expression data from the Cancer Genome Atlas (TCGA-PRAD) (20). PDCD4 protein levels were inversely correlated with miR-21 expression (Spearman r = −0.18, p < 0.001) in 350 available matched patient samples (Fig. 3A). The levels of PDCD4 mRNA were also inversely correlated with miR-21 expression (Spearman r = −0.21, p < 0.0001) in the 546 available matched patient samples (Fig. 3B). These correlations were similar in strength and significance when compared to those found for PDCD4 mRNA and PDCD4 protein (Spearman r = 0.24, p < 0.0001) (Fig. 3C). We then examined the association of miR-21 and PDCD4 gene expression with radical prostatectomy Gleason Score. PDCD4 mRNA and protein levels were significantly decreased in higher Gleason score tumors (Fig. 3D-E, One-way ANOVA, p < 0.05), while the levels of miR-21 were significantly increased (Fig 3F, p < 0.0001).
Figure 3. PDCD4 and miR-21 expression are inversely correlated and associated with aggressive prostate cancer.
Analysis of miR-21 miRNA, PDCD4 mRNA, and PDCD4 protein expression from the TCGA-PRAD. Spearman correlation analysis of normalized of (A) miR-21 (RPM) and PDCD4 protein (RPPA) expression in 350 matching TCGA-PRAD samples, (B) miR-21 (RPM) and PDCD4 mRNA (RPM) expression in 546 matching TCGA-PRAD samples, and (C) PDCD4 mRNA and protein in 351 matching TCGA-PRAD samples. Association of (D) PDCD4 protein, (E) PDCD4 mRNA, and (F) miR-21 with radical prostatectomy Gleason Score in TCGA-PRAD tissue samples (One-Way ANOVA).
miR-21 induces androgen-dependent and androgen-independent growth through PDCD4 knockdown
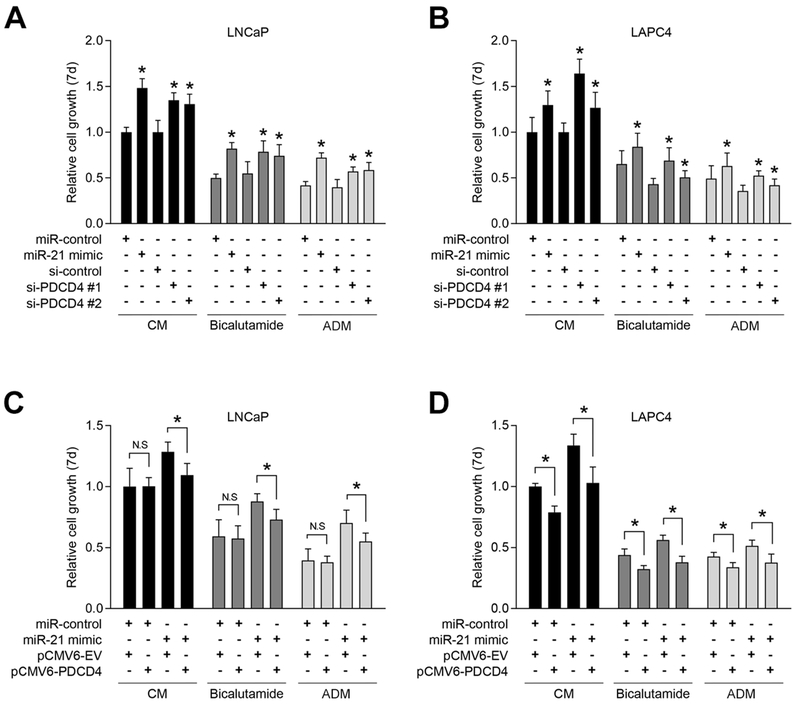
To further study role of miR-21 and PDCD4 in PCa proliferation and castration resistance, we applied the same androgen-dependent cell lines, LNCaP and LAPC4, and a second cell proliferation assay, the MTS assay. We inhibited PDCD4 expression with two separate siRNAs, and we over-expressed PDCD4 with the pCMV6-PDCD4 vector. Western blotting verified PDCD4 knockdown and over-expression in both cell models (Supplementary Fig. S3). PDCD4 knockdown by both siRNAs significantly enhanced androgen-dependent growth in CM, when compared to si-control, in both cell lines (Fig. 4A-B, CM, black bars). The level of growth induced by both PDCD4 siRNAs was comparable to that induced by miR-21 mimics. To study androgen-independent growth, cells were cultured in CM supplemented with Bicalutamide (Fig. 4, dark grey bars) or in ADM (Fig. 4, light grey bars). Androgen deprivation, by Bicalutamide or ADM, significantly decreased cell growth when compared to cells grown in CM. Notably, PDCD4 knockdown drove androgen-independent cell growth in both cell line models, either when AR was directly inhibited with Bicalutamide (Fig. 4A-B, dark grey bars) or when cells were grown in ADM (Fig. 4A-B, light grey bars). The overall level of androgen-independent growth was similar in miR-21 mimic and PDCD4 siRNA transfected cells. These results are consistent with the original siRNA screening experiments (Fig. 1B-C), and they support that PDCD4 knockdown is sufficient to drive androgen-independent PCa growth.
Figure 4. PDCD4 knockdown induces androgen-dependent and androgen-independent prostate cancer growth.
A and B, PDCD4-knockdown-induced androgen-dependent and androgen-independent cell growth. LNCaP or LAPC4 cells were transiently transfected with 20 nM of miR-control or miR-21 mimic, or 20 nM of si-control or PDCD4 siRNAs. Forty-eight hours after transfection, cells were grown in CM, CM + 10 μM Bicalutamide, or ADM for 7 days. Cell viability was evaluated by MTS assay and reported relative to miR-control signal in CM. Mean + S.E. from twelve independent measurements. *, p < 0.05, relative to miR-control or si-control. C and D, Cells were co-transfected with miR-control or miR-21 mimic and pCMV6-empty vector (EV) or pCMV6-PDCD4. After 48 h, cells were grown in CM, CM + 10 μM Bicalutamide, or ADM for 7 days. Cell viability was evaluated by MTS assay and reported relative to miR-control signal in CM. Mean + S.E. from twelve independent measurements. *, p < 0.05.
To determine whether miR-21 functioned through PDCD4 knockdown, we applied a PDCD4 expression vector that lacked the natural 3’UTR and corresponding miR-21 binding sites. The empty vector, pCMV6-EV, was applied as a negative control. Results demonstrate that PDCD4 over-expression significantly blocked miR-21-induced cell growth in androgen-containing CM (Fig. 4C-D, black bars). Importantly, PDCD4 over-expression also blocked the androgen-independent growth driven by miR-21 in Bicalutamide treated cells (Fig. 4C-D, dark grey bars) and in cells grown in ADM (Fig. 4C-D, light grey bars). In the absence of miR-21 over-expression (miR-control), elevated PDCD4 expression inhibited LAPC4, but not in LNCaP, cell growth under all treatment conditions (Fig. 4C-D). This decrease in LAPC4 cell growth was associated with an increase in Annexin V positive cells, which was not similarly found in LNCaP cells (Supplementary Fig. S4).
PDCD4 knockdown induces cell cycle progression and reduces apoptosis in a cell-specific manner
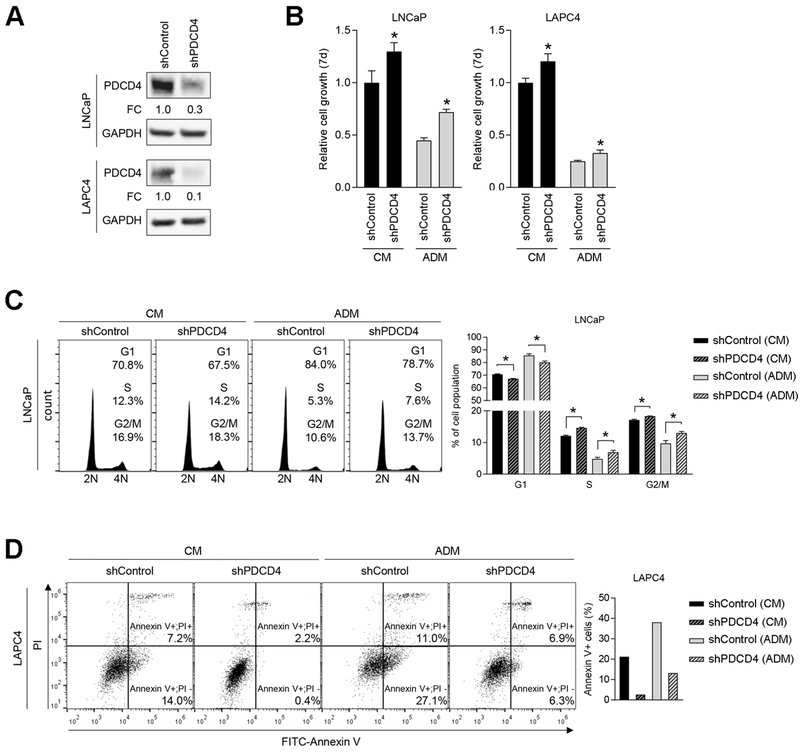
To expand upon the mechanism of PDCD4-mediated growth suppression, we generated two stable PDCD4 knockdown cell lines, LNCaP-shPDCD4 and LAPC4-shPDCD4, and we evaluated changes in cell proliferation and apoptosis. Corresponding negative control cell lines were generated with the shControl vector. Western blotting demonstrated shRNA-mediated PDCD4 knockdown of 70-90% (Fig. 5A). Both shPDCD4 cell lines demonstrated enhanced androgen-dependent growth and androgen-independent growth when compared to shControl cells (Fig. 5B). In cell cycle analyses, LNCaP-shPDCD4 cells exhibited a lower percentage of cells in G1 phase and a larger percentage in S and G2/M phases, when compared to shControl cells, in both CM and ADM conditions (Fig. 5C), suggesting that PDCD4 functions through cell cycle regulation. However, surprisingly, LAPC4-shPDCD4 cells did not show the same changes in cell cycle progression when compared to shControl cells (Supplementary Fig. S5A). Instead, LAPC4-shPDCD4 showed a significant decreased in the level of apoptosis when grown in CM and AD and compared to shControl cells (Fig. 5D). This anti-apoptotic effect of PDCD4 was not found in LNCaP-shPDCD4 cells (Supplementary Fig. S5B). Collectively, these results indicate a cell-specific response to PDCD4 knockdown, which includes enhanced proliferation and reduced apoptosis.
Figure 5. PDCD4 knockdown induces cell cycle progression and inhibits apoptosis in a cell specific manner.
A, Stable PDCD4 knockdown lines. LNCaP or LAPC4-shControl and -shPDCD4 cells were generated by lentiviral transduction. PDCD4 protein expression measured by WB, normalized to GAPDH. FC, Fold Change relative to shControl. B, LNCaP (left panel) or LAPC4 (right panel) cell growth in CM and ADM as measured by MTS assay (7 days). Mean + S.E. from twelve independent measurements. *, p < 0.05, relative to shControl. C, Cell cycle analysis in CM or ADM by flow cytometry, 72 h. Left figure represents one of three independent replicates and right graph shows combined three biologic replicates. Mean + S.E. (n = 3). *, p < 0.05. D, Apoptotic analysis by Annexin V staining and flow cytometry after 72 h growth in CM or ADM. Left figure represents one of three independent replicates and right graph indicates total percentage of Annexin V+ cell population.
PDCD4 may regulate cell cycle progression by inhibiting eIF4A, an RNA helicase and translation initiation factor. PDCD4 knockdown induced the expression of c-Myc and Cyclin E1, two eIF4A regulated transcripts associated with cell cycle progression (28,29), in LNCaP-shPDCD4 cells, but not LAPC4-shPDCD4 cells (Supplementary Fig. S5C). Further experiments are required to determine if eIF4A is a key downstream target of miR-21 and PDCD4 in PCa cells.
PDCD4 knockdown enhances PCa tumorigenesis, tumor growth rate, and imparts castration resistance
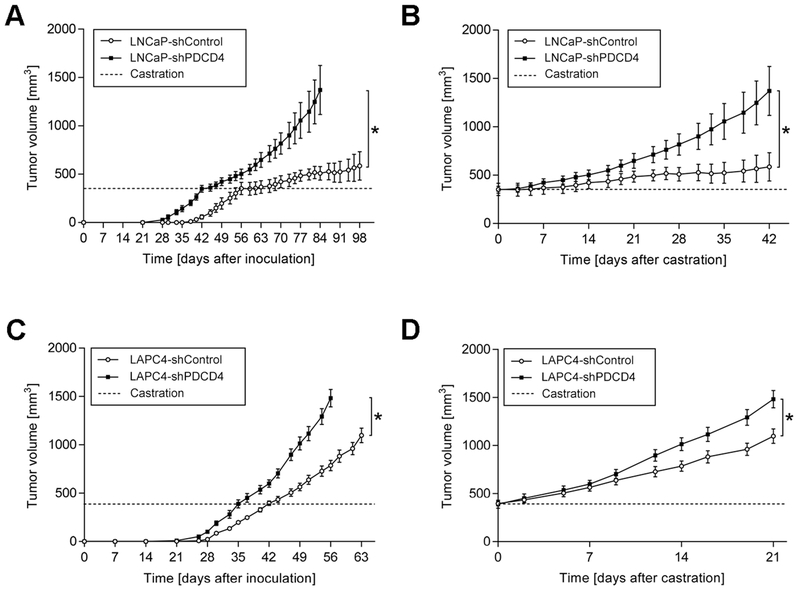
To study the impact of PDCD4 knockdown on PCa cell growth in vivo, we developed subcutaneous xenograft tumors using shPDCD4 and shControl cells in male athymic nude mice. Strikingly, the tumor take rate of LNCaP-shPDCD4 cells was considerably higher, when compared to LNCaP-shControl cells (90 % versus 60 %, respectively). The onset of palpable tumor detection was also 7 days earlier for LNCaP-shPDCD4 cells, when compared to LNCaP-shControl tumors (Fig. 6A). Further, LNCaP-shPDCD4 tumors grew at a faster rate. When tumors reached an average volume of 350 mm3, mice were castrated by bilateral orchiectomy (Fig. 6, dashed line). Castration markedly reduced LNCaP-shControl tumor growth, whereas LNCaP-shPDCD4 tumors continued to grow rapidly. This castration resistant growth is most apparent when tumor growth is normalized to the time of orchiectomy (Fig. 6B). In the stable LAPC4 cell line models, shPDCD4 tumors similarly showed earlier onset of tumorigenesis and enhanced tumor growth rate, when compared to shControl tumors (Fig. 6C). The tumor take rate was also higher in LAPC4-shPDCD4 cells, when compared to LAPC4-shControl cells (70 % versus 50 %, respectively). The impact of castration was less dramatic in LAPC4 cells, when compared to LNCaP cells, as has been experienced by others (30). Nonetheless, LAPC4-shPDCD4 tumors grew significantly faster than LAPC4-shControl tumors after castration (Fig. 6D). Collectively, these results demonstrate that reduced PDCD4 expression promotes PCa tumorigenesis, tumor growth, and castration resistant tumor growth.
Figure 6. PDCD4 knockdown enhances tumorigenesis, increases tumor growth rate, and imparts castration resistance in vivo.
A, Subcutaneous xenograft growth of LNCaP-shControl or LNCaP-shPDCD4 over time, initiating from the time of injection. Dashed line indicates castration when tumors reached ~350 mm3 volume. B, Tumor growth over time, normalized to the time of castration. Mean ± S.E. (LNCaP-shControl, n = 6; LNCaP-shPDCD4, n = 9). *, p < 0.05; Two-Way ANOVA. C, Subcutaneous xenograft growth of LAPC4-shControl or LAPC4-shPDCD4 over time, initiating with the time of injection. Dashed line indicates castration when tumors reached ~380 mm3 volume. D, Tumor growth over time, normalized to the time of castration. Mean ± S.E. (LAPC4-shControl, n = 5; LAPC4-shPDCD4, n = 7). *, p < 0.05; Two-Way ANOVA.
DISCUSSION
Despite the recent development of several new life-prolonging therapies (6), CRPC remains an incurable disease. Improved CRPC treatment may come through a better understanding of the mechanisms capable of driving CRPC growth. Apart from the rare neuroendocrine phenotype, CRPC can be divided into two primary groups, those that are AR-dependent and those that are AR-independent. AR-dependent CRPC can develop through AR gene mutation, AR gene amplification, alternative AR splicing, aberrant AR co-factor activity, or anomalous androgen production (4). Seeking pathways downstream of AR that are capable of causing castration resistant growth, we identified miR-21 (16). The activated AR directly induces the expression of the primary miR-21 transcript, pri-miR-21, by binding to the miPPR21 promoter as demonstrated by chromatin immunoprecipitation experiments and reporter assays (16,27). Several AR-independent pathways, including AP-1, RAS, IL-6/STAT3, chromatin modification, and alternative polyadenylation of VMP1, can also induce the expression of miR-21 (12–14,31). Thus, there is potential for elevated miR-21 expression through AR-dependent or AR-independent signaling. Here we sought to characterize pathways downstream of miR-21 that are capable of driving castration resistant PCa growth. Our results implicate PDCD4 suppression as a novel mechanism for enhanced PCa tumorigenesis, tumor growth rate, and castration resistance.
PDCD4 (Programmed Cell Death 4) originally acquired its name because it was found to be induced upon programmed cell death (32). However, PDCD4 expression and function are not restricted to apoptosis. One important role of PDCD4 is as a post-transcriptional regulator of cap-dependent protein translation. PDCD4 binds and inhibits the eukaryotic translation initiation factor and RNA helicase, eIF4A (33–35). It has been proposed that PDCD4 regulates gene transcripts with complex 5’UTRs through two possible mechanism, by directly binding and sequestering eIF4A, and separately, by binding nuclear mRNAs and preventing their transfer to the cytoplasm (36). Cap-dependent translation initiation, through eIF4A, regulates the expression of several cancer-associated genes, including c-Myc, cyclins, and PARP (28,37,38). With this broad potential for gene regulation, PDCD4 inhibition can produce different phenotypic changes in different cell types (39). Consistent with this, we found PDCD4 knockdown to induce proliferation in one model, LNCaP, and to inhibit apoptosis in another, LAPC4. In LNCaP cells, PDCD4 knockdown induced c-Myc and Cyclin E1 expression, possibly through enhanced eIF4A mediated cap-dependent translation. Often, tumor suppressive pathways can be flexible and dependent upon cell type, differentiation state, stress conditions, and active signaling pathways (40). While LNCaP and LAPC4 cells are both AR positive and androgen-sensitive, they differ in p53 and PTEN gene status (41,42). Further experimentation is needed to define the direct mechanism of PDCD4 in these and other PCa cell lines.
Accumulating evidence suggests that PDCD4 is a true tumor suppressor. PDCD4-knockout mice develop spontaneous lymphomas (43). Transgenic mice over-expressing PDCD4 produce significantly less tumors in the DMBA/TPA skin carcinogenesis model (44). Here, for the first time, we reveal the role of PDCD4 in androgen-dependent PCa cells and tumors. We find that reduced PDCD4 expression causes enhanced PCa tumorigenesis, tumor growth rate, and castration resistance.
The present study is also the first to report PDCD4 as an androgen suppressed protein, which occurs at least partially through miR-21 expression. Mitogen induced S6K1 signaling and βTRCP-mediated proteasome degradation can also lead to the rapid loss of PDCD4 protein levels (45). Our data suggest that miR-21 is a dominant inhibitor of androgen-induced PDCD4 protein loss, because miR-21 inhibition rescued PDCD4 expression to 80-90% of basal levels following androgen treatment. It is notable that there is potential for crosstalk between these two pathways, because PTEN is a known miR-21 target, and PTEN suppresses the PI3K/mTOR/S6K pathway (46). However, a large proportion of our studies were performed in the LNCaP cell line model, which is PTEN deficient. Additional studies are needed to distinguish the roles of AR, miR-21, PTEN, and PI3K/mTOR/S6K signaling in PDCD4 protein regulation in PCa.
Previous studies have investigated the association of miR-21, and PDCD4, with aggressive features of localized PCa. Elevated miR-21 expression is associated with poor biochemical recurrence-free survival and PCa progression (47,48); however, miR-21 levels alone may not provide substantially greater prognostic value beyond established clinical parameters such as Gleason Score or disease stage (49). Reduced PDCD4 protein expression has similarly been associated with PCa progression and aggressive pathologic features (50). Our analyses of the TCGA-PRAD dataset provide a unique insight into miR-21, PDCD4 mRNA, and PDCD4 protein levels in the same human tumor samples. These results show significant, but weak, inverse correlation between miR-21 expression and PDCD4 mRNA and protein levels. There is also a clear association between elevated miR-21 miRNA, and reduced PDCD4 protein and mRNA, and advanced grade disease in TCGA samples. These trends are consistent with the tumor suppressive nature of PDCD4 in LNCaP and LAPC4 cells. It is notable that PDCD4 expression values overlap in some samples among these disease states. Further detailed studies are needed to determine whether PDCD4 expression, or cellular localization, could provide new prognostic value for patients with localized disease.
In conclusion, we reveal PDCD4 as an androgen-suppressed protein. Androgen signaling inhibits PDCD4 expression through miR-21, and loss of PDCD4 expression induces PCa proliferation and reduces cellular apoptosis. Loss of PDCD4 expression is sufficient to enhance tumorigenesis and to impart castration resistance in human tumor xenograft models. The miR-21/PDCD4 signaling pathway may represent a novel target for assessing or treating locally aggressive PCa or metastatic CRPC.
Supplementary Material
Implications:
This study provides the first evidence that PDCD4 is an androgen-suppressed protein capable of regulating PCa cell proliferation, apoptosis, and castration resistance. These results uncover miR-21 and PDCD4 regulated pathways as potential new targets for castration resistant prostate cancer.
Acknowledgments
This work was supported by the National Institutes of Health (1R01CA143299, 5P50CA058236, P30CA006973), the Department of Defense (PC073745), and the Patrick C. Walsh Prostate Cancer Research Fund. We thank Judit Ribas and Luzia Brander for analyses of additional miR-21 target genes.
Financial Support: This work was supported by research grants R01CA143299, P50CA058236, P30CA006973 (SEL) from the National Institutes of Health, W81XWH-17-1-0581 (SEL) from the Department of Defense, and the Patrick C. Walsh Prostate Cancer Research Fund.
Footnotes
Conflict of interest
The authors declare no conflicts of interest.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: a cancer journal for clinicians 2015;65(2):87–108 doi 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA: a cancer journal for clinicians 2018;68(1):7–30 doi 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 3.Wong YN, Ferraldeschi R, Attard G, de Bono J. Evolution of androgen receptor targeted therapy for advanced prostate cancer. Nature reviews Clinical oncology 2014;11(6):365–76 doi 10.1038/nrclinonc.2014.72. [DOI] [PubMed] [Google Scholar]
- 4.Debes JD, Tindall DJ. Mechanisms of androgen-refractory prostate cancer. The New England journal of medicine 2004;351(15):1488–90 doi 10.1056/NEJMp048178. [DOI] [PubMed] [Google Scholar]
- 5.Lu J, Van der Steen T, Tindall DJ. Are androgen receptor variants a substitute for the full-length receptor? Nature reviews Urology 2015;12(3):137–44 doi 10.1038/nrurol.2015.13. [DOI] [PubMed] [Google Scholar]
- 6.Hamid AA, Sweeney CJ. Prostate cancer: A new standard-of-care for advanced-stage disease. Nature reviews Clinical oncology 2017;14(10):592–3 doi 10.1038/nrclinonc.2017.120. [DOI] [PubMed] [Google Scholar]
- 7.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005;120(1):15–20 doi 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 8.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature 2010;467(7311):86–90 doi 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 9.Volinia S, Galasso M, Costinean S, Tagliavini L, Gamberoni G, Drusco A, et al. Reprogramming of miRNA networks in cancer and leukemia. Genome research 2010;20(5):589–99 doi 10.1101/gr.098046.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hart M, Nolte E, Wach S, Szczyrba J, Taubert H, Rau TT, et al. Comparative microRNA profiling of prostate carcinomas with increasing tumor stage by deep sequencing. Molecular cancer research : MCR 2014;12(2):250–63 doi 10.1158/1541-7786.MCR-13-0230. [DOI] [PubMed] [Google Scholar]
- 11.Jalava SE, Urbanucci A, Latonen L, Waltering KK, Sahu B, Janne OA, et al. Androgen-regulated miR-32 targets BTG2 and is overexpressed in castration-resistant prostate cancer. Oncogene 2012;31(41):4460–71 doi 10.1038/onc.2011.624. [DOI] [PubMed] [Google Scholar]
- 12.Fujita S, Ito T, Mizutani T, Minoguchi S, Yamamichi N, Sakurai K, et al. miR-21 Gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. Journal of molecular biology 2008;378(3):492–504 doi 10.1016/j.jmb.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 13.Talotta F, Cimmino A, Matarazzo MR, Casalino L, De Vita G, D’Esposito M, et al. An autoregulatory loop mediated by miR-21 and PDCD4 controls the AP-1 activity in RAS transformation. Oncogene 2009;28(1):73–84 doi 10.1038/onc.2008.370. [DOI] [PubMed] [Google Scholar]
- 14.Loffler D, Brocke-Heidrich K, Pfeifer G, Stocsits C, Hackermuller J, Kretzschmar AK, et al. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood 2007;110(4):1330–3 doi 10.1182/blood-2007-03-081133. [DOI] [PubMed] [Google Scholar]
- 15.Zhong X, Chung AC, Chen HY, Meng XM, Lan HY. Smad3-mediated upregulation of miR-21 promotes renal fibrosis. Journal of the American Society of Nephrology : JASN 2011;22(9):1668–81 doi 10.1681/ASN.2010111168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ribas J, Ni X, Haffner M, Wentzel EA, Salmasi AH, Chowdhury WH, et al. miR-21: an androgen receptor-regulated microRNA that promotes hormone-dependent and hormone-independent prostate cancer growth. Cancer research 2009;69(18):7165–9 doi 10.1158/0008-5472.CAN-09-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 2008;27(15):2128–36 doi 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 18.Lu Z, Liu M, Stribinskis V, Klinge CM, Ramos KS, Colburn NH, et al. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene 2008;27(31):4373–9 doi 10.1038/onc.2008.72. [DOI] [PubMed] [Google Scholar]
- 19.Lupold SE, Johnson T, Chowdhury WH, Rodriguez R. A real time Metridia luciferase based non-invasive reporter assay of mammalian cell viability and cytotoxicity via the beta-actin promoter and enhancer. PloS one 2012;7(5):e36535 doi 10.1371/journal.pone.0036535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cancer Genome Atlas Research N. The Molecular Taxonomy of Primary Prostate Cancer. Cell 2015;163(4):1011–25 doi 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan LX, Wu QN, Zhang Y, Li YY, Liao DZ, Hou JH, et al. Knockdown of miR-21 in human breast cancer cell lines inhibits proliferation, in vitro migration and in vivo tumor growth. Breast cancer research : BCR 2011;13(1):R2 doi 10.1186/bcr2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel-Duby R, van Rooij E, et al. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer cell 2010;18(3):282–93 doi 10.1016/j.ccr.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu G, Zhang Y, Wei J, Jia W, Ge Z, Zhang Z, et al. MicroRNA-21 promotes hepatocellular carcinoma HepG2 cell proliferation through repression of mitogen-activated protein kinase-kinase 3. BMC cancer 2013;13:469 doi 10.1186/1471-2407-13-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell research 2008;18(3):350–9 doi 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 25.Zhong Z, Dong Z, Yang L, Gong Z. miR-21 induces cell cycle at S phase and modulates cell proliferation by down-regulating hMSH2 in lung cancer. Journal of cancer research and clinical oncology 2012;138(10):1781–8 doi 10.1007/s00432-012-1287-y. [DOI] [PubMed] [Google Scholar]
- 26.Han L, Yue X, Zhou X, Lan FM, You G, Zhang W, et al. MicroRNA-21 expression is regulated by beta-catenin/STAT3 pathway and promotes glioma cell invasion by direct targeting RECK. CNS neuroscience & therapeutics 2012;18(7):573–83 doi 10.1111/j.1755-5949.2012.00344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ribas J, Lupold SE. The transcriptional regulation of miR-21, its multiple transcripts, and their implication in prostate cancer. Cell cycle 2010;9(5):923–9 doi 10.4161/cc.9.5.10930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiegering A, Uthe FW, Jamieson T, Ruoss Y, Huttenrauch M, Kuspert M, et al. Targeting Translation Initiation Bypasses Signaling Crosstalk Mechanisms That Maintain High MYC Levels in Colorectal Cancer. Cancer discovery 2015;5(7):768–81 doi 10.1158/2159-8290.CD-14-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oblinger JL, Burns SS, Huang J, Pan L, Ren Y, Shen R, et al. Overexpression of eIF4F components in meningiomas and suppression of meningioma cell growth by inhibiting translation initiation. Experimental neurology 2018;299(Pt B):299–307 doi 10.1016/j.expneurol.2017.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang KH, Li R, Papari-Zareei M, Watumull L, Zhao YD, Auchus RJ, et al. Dihydrotestosterone synthesis bypasses testosterone to drive castration-resistant prostate cancer. Proceedings of the National Academy of Sciences of the United States of America 2011;108(33):13728–33 doi 10.1073/pnas.1107898108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ribas J, Ni X, Castanares M, Liu MM, Esopi D, Yegnasubramanian S, et al. A novel source for miR-21 expression through the alternative polyadenylation of VMP1 gene transcripts. Nucleic acids research 2012;40(14):6821–33 doi 10.1093/nar/gks308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shibahara K, Asano M, Ishida Y, Aoki T, Koike T, Honjo T. Isolation of a novel mouse gene MA-3 that is induced upon programmed cell death. Gene 1995;166(2):297–301. [DOI] [PubMed] [Google Scholar]
- 33.Lankat-Buttgereit B, Goke R. The tumour suppressor Pdcd4: recent advances in the elucidation of function and regulation. Biology of the cell 2009;101(6):309–17 doi 10.1042/BC20080191. [DOI] [PubMed] [Google Scholar]
- 34.Yang HS, Cho MH, Zakowicz H, Hegamyer G, Sonenberg N, Colburn NH. A novel function of the MA-3 domains in transformation and translation suppressor Pdcd4 is essential for its binding to eukaryotic translation initiation factor 4A. Molecular and cellular biology 2004;24(9):3894–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang HS, Jansen AP, Komar AA, Zheng X, Merrick WC, Costes S, et al. The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Molecular and cellular biology 2003;23(1):26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vikhreva PN, Kalinichenko SV, Korobko IV. Programmed cell death 4 mechanism of action: The model to be updated? Cell cycle 2017;16(19):1761–4 doi 10.1080/15384101.2017.1371881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Modelska A, Turro E, Russell R, Beaton J, Sbarrato T, Spriggs K, et al. The malignant phenotype in breast cancer is driven by eIF4A1-mediated changes in the translational landscape. Cell death & disease 2015;6:e1603 doi 10.1038/cddis.2014.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rubio CA, Weisburd B, Holderfield M, Arias C, Fang E, DeRisi JL, et al. Transcriptome-wide characterization of the eIF4A signature highlights plasticity in translation regulation. Genome biology 2014;15(10):476 doi 10.1186/s13059-014-0476-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lankat-Buttgereit B, Lenschen B, Schmidt H, Goke R. The action of Pdcd4 may be cell type specific: evidence that reduction of dUTPase levels might contribute to its tumor suppressor activity in Bon-1 cells. Apoptosis : an international journal on programmed cell death 2008;13(1):157–64 doi 10.1007/s10495-007-0153-x. [DOI] [PubMed] [Google Scholar]
- 40.Kastenhuber ER, Lowe SW. Putting p53 in Context. Cell 2017;170(6):1062–78 doi 10.1016/j.cell.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Bokhoven A, Varella-Garcia M, Korch C, Johannes WU, Smith EE, Miller HL, et al. Molecular characterization of human prostate carcinoma cell lines. The Prostate 2003;57(3):205–25 doi 10.1002/pros.10290. [DOI] [PubMed] [Google Scholar]
- 42.Liu P, Li S, Gan L, Kao TP, Huang H. A transcription-independent function of FOXO1 in inhibition of androgen-independent activation of the androgen receptor in prostate cancer cells. Cancer research 2008;68(24):10290–9 doi 10.1158/0008-5472.CAN-08-2038. [DOI] [PubMed] [Google Scholar]
- 43.Hilliard A, Hilliard B, Zheng SJ, Sun H, Miwa T, Song W, et al. Translational regulation of autoimmune inflammation and lymphoma genesis by programmed cell death 4. Journal of immunology 2006;177(11):8095–102. [DOI] [PubMed] [Google Scholar]
- 44.Jansen AP, Camalier CE, Colburn NH. Epidermal expression of the translation inhibitor programmed cell death 4 suppresses tumorigenesis. Cancer research 2005;65(14):6034–41 doi 10.1158/0008-5472.CAN-04-2119. [DOI] [PubMed] [Google Scholar]
- 45.Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE, Pagano M. S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science 2006;314(5798):467–71 doi 10.1126/science.1130276. [DOI] [PubMed] [Google Scholar]
- 46.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 2007;133(2):647–58 doi 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guan Y, Wu Y, Liu Y, Ni J, Nong S. Association of microRNA-21 expression with clinicopathological characteristics and the risk of progression in advanced prostate cancer patients receiving androgen deprivation therapy. The Prostate 2016;76(11):986–93 doi 10.1002/pros.23187. [DOI] [PubMed] [Google Scholar]
- 48.Li T, Li RS, Li YH, Zhong S, Chen YY, Zhang CM, et al. miR-21 as an independent biochemical recurrence predictor and potential therapeutic target for prostate cancer. The Journal of urology 2012;187(4):1466–72 doi 10.1016/j.juro.2011.11.082. [DOI] [PubMed] [Google Scholar]
- 49.Zheng Q, Peskoe SB, Ribas J, Rafiqi F, Kudrolli T, Meeker AK, et al. Investigation of miR-21, miR-141, and miR-221 expression levels in prostate adenocarcinoma for associated risk of recurrence after radical prostatectomy. The Prostate 2014;74(16):1655–62 doi 10.1002/pros.22883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li CR, Su JJ, Wang WY, Lee MT, Wang TY, Jiang KY, et al. Molecular profiling of prostatic acinar morphogenesis identifies PDCD4 and KLF6 as tissue architecture-specific prognostic markers in prostate cancer. The American journal of pathology 2013;182(2):363–74 doi 10.1016/j.ajpath.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.