Abstract
DNA methylation, catalyzed by DNA methyltransferases (DNMTs), is an epigenetic mark that needs to be faithfully replicated during mitosis in order to maintain cell phenotype during successive cell divisions. This epigenetic mark is located on the 5′-carbon of the cytosine mainly within cytosine–phosphate–guanine (CpG) dinucleotides. DNA methylation is asymmetrically positioned on both DNA strands, temporarily generating a hemi-methylated state after DNA replication. Hemi-methylation is a particular status of DNA that is recognized by ubiquitin-like containing plant homeodomain (PHD) and really interesting new gene (RING) finger domains 1 (UHRF1) through its SET- (Su(var)3-9, Enhancer-of-zeste and Trithorax) and RING-associated (SRA) domain. This interaction is considered to be involved in the recruitment of DNMT1 to chromatin in order to methylate the adequate cytosine on the newly synthetized DNA strand. The UHRF1/DNMT1 tandem plays a pivotal role in the inheritance of DNA methylation patterns, but the fine-tuning mechanism remains a mystery. Indeed, because DNMT1 experiences difficulties in finding the cytosine to be methylated, it requires the help of a guide, i.e., of UHRF1, which exhibits higher affinity for hemi-methylated DNA vs. non-methylated DNA. Two models of the UHRF1/DNMT1 dialogue were suggested to explain how DNMT1 is recruited to chromatin: (i) an indirect communication via histone H3 ubiquitination, and (ii) a direct interaction of UHRF1 with DNMT1. In the present review, these two models are discussed, and we try to show that they are compatible with each other.
Keywords: cell identity, DNA methylation, DNMT1, epigenetics, gene expression, UHRF1
1. Introduction
1.1. DNA Methylation Patterns: Layers of Epigenomes
DNA methylation mainly occurs on cytosine–phosphate–guanine (CpG) dinucleotides, and almost 70–80% of all CpG dinucleotides (~3 × 107) are methylated in human cell genomes [1,2]. DNA methylation can also occur at non-CpG, including cytosine–phosphate–adenine (CpA), cytosine–phosphate–thymine (CpT), and cytosine–phosphate–cytosine (CpC), with the first one being the most frequent [3]. The role of DNA methylation is still a matter of debate [3]. Considering that de novo DNA methyltransferases (DNMTs), such as DNMT3A and DNMT3B, catalyze methylation at non-CpG sites, we do not extend this review to non-CpG methylation. Although the role of DNA methylation was reviewed elsewhere, before going deeper into the insights of the mechanism of DNA methylation pattern duplication, a few reminders on the location of the methylated CpG sites are made hereafter.
Methylated CpG dinucleotides are preferentially localized within repetitive elements (SINEs; Short Interspersed Nuclear Elements, LINEs; Long INterspersed Nuclear Elements, and LTR; Long Terminal Repeat), centromeres, and coding regions of functional genes (gene bodies) [4]. Methylation of CpG dinucleotides in these areas guarantees genomic stability and prevents aberrant initiation of transcription [5,6]. The primary well-recognized role of DNA methylation lies in transcriptional repression of retrotransposons, X chromosome inactivation, and mono-allelic expression of imprinted genes [7]. Therefore, the methylation of DNA, along with enhancers and gene bodies [8,9,10], is involved in gene expression regulation. Non-methylated CpG dinucleotides are frequently observed in promoter regions of active genes where they form “CpG islands (CGIs)”. CpG islands are regions greater than 550 bp of DNA with a GC content greater than 50%, and an observed CpG/expected CpG ratio greater than 0.6 [11]. Approximately, 70% of human gene promoters contain CGIs, usually unmethylated in normal cells, thus allowing transcription of the corresponding gene. During development and differentiation, around 6% of them undergo methylation in a tissue-specific manner, thereby contributing to the occurrence of different phenotypes [5,11]. Irregularities in methylation pattern, especially in promoters, can lead to profound abnormalities including malignant transformation of cells [4]. In cancers, two opposite patterns of DNA methylation are observed and contribute together to tumorigenesis. Firstly, CpG islands in promoter regions of various tumor suppressor genes (TSGs), such as p16INK4A, p73, BRCA1, and TIMP3, are hypermethylated in cancer, inhibiting their expression and, thus, leading to uncontrolled proliferation of cancer cells [12,13,14,15]. Nonetheless, establishment of this methylation pattern is not yet deciphered. Secondly, a global hypomethylation is observed in cancer cells, which promotes the expression of oncogenes and aggravates tumorigenesis by inducing genomic instability [16,17]. The origin of these two upheavals in DNA methylation patterns is not yet fully elucidated, but might find a beginning of an explanation in the altered communication between ubiquitin-like containing plant homeodomain (PHD) and really interesting new gene (RING) finger domains 1 (UHRF1) and DNA methyltransferase 1 (DNMT1) [18,19], which is the matter of the present review. However, what is clear is that these DNA methylation patterns are faithfully transmitted from a mother cancer cell to the two daughter cancer cells with a phenotype preserved almost to infinity. Considering that cancer cells usually divide faster than normal cells and that they have a more stable phenotype throughout successive mitosis, especially under in vitro culture conditions [20], one might imagine that the DNMT1/UHRF1 tandem is more efficient in cancer cells than non-cancerous cells.
Efforts over the past decade shed light onto the dynamics and the molecular mechanisms underlying DNA methylation pattern duplication occurring after DNA replication. One important breakthrough was achieved by the discovery of the UHRF1/DNMT1 network.
1.2. The Role of UHRF1/DNMT1 Tandem
Chromosomal mapping of human UHRF1 and DNMT1 genes both at 19p13, more specifically at 19p13.3 and 19p13.2, respectively, [21] and genome sequencing revealed that they are separated by about 50 Mb, i.e., 50 centimorgans (personal observations). It is worth noting that the tissue-specific expression of UHRF1 and DNMT1 is tightly linked, suggesting that they need each other to exert their role. Indeed, for both genes, the most elevated expressions were found in the appendix, bone marrow, lymph node, and testis [22,23]. In contrast, highly differentiated tissues, such as the heart, liver, pancreas, prostate, and salivary glands, were among the tissues that express the lowest levels of DNMT1 and UHRF1 messenger RNAs (mRNAs) [22,23]. This may explain that UHRF1−/− phenocopies DNMT1−/− [24,25], that DNMT−/− is embryonic lethal at day 9 [26], and that UHRF1−/− is mid-gestational lethal [27], suggesting that both genes have interdependent roles in DNA methylation maintenance. This also suggests that both genes might have similar pathways of regulation. Pharmacological or pathological regulation of the expression of UHRF1 and DNMT1 genes was extensively reviewed elsewhere [22,28,29,30,31,32,33,34,35,36,37]. Briefly, one interesting point with deep impact is that downregulation of UHRF1 and/or DNMT1 always allows re-expression or enhanced expression of a large number of tumor suppressor genes, including RB1, p16INK4A, CDH13, SHP1, SOCS3, 3OST2, BRCA1, CDX2, RUNX3, FOXO4, PPARG, PML, MEG3, HHIP, IGFBP3, SFRP1, and 14-3-3σ [29,34,35,38,39,40,41,42,43,44,45]. Also of note, it was shown that UHRF1 is involved in epigenetic silencing of KiSS1, a metastasis suppressor gene [46]. The mechanism of the re-expression of tumor suppressor genes following decreased expression of DNMT1 and UHRF1 remains elusive. Indeed, it is clear that these two proteins are involved in the maintenance of hypermethylation of promoters, but how they are demethylated remains a mystery. A passive demethylation, via a downregulation of UHRF1, through successive mitosis appears unlikely, as UHRF1 is indispensable for cell proliferation [22,47].
For global DNA hypomethylation, a diminished interaction was suggested between UHRF1 and DNMT1 [18,19], although UHRF1 expression is enhanced in all cancers so far investigated [29,34,35,36,48]. However, a ubiquitin-dependent degradation of DNMT3A induced by UHRF1 and/or UHRF2 might also be involved [49]. Indeed, considering that DNMT3A is involved in de novo DNA methylation, an increase of UHRF1 through the targeting of DNMT3A also likely contributes to the global DNA hypomethylation in cancer cells [49].
The maintenance of DNA methylation at the replication fork is believed to be ensured by the DNMT1/PCNA (Proliferating Cell Nuclear Antigen) tandem [50,51,52,53]; however, surprisingly, its disruption exerts little effect on genomic DNA methylation in contrast to that of the UHRF1/DNMT1 tandem, which induces massive DNA hypomethylation [18]. Furthermore, the occurrence of a disruption in the oncogenic process was extended to the UHRF1/DNMT1/PCNA complex [19]. Complementary to this event, it was demonstrated that UHRF1 overexpression drives DNA hypomethylation by delocalizing DNMT1 [54], which further supports that abnormal cooperation within the UHRF1/DNMT1 tandem may be one of the first steps of tumorigenesis onset. Indeed, the de novo methylation of genes frequently observed in cancers could be catalyzed by DNMT1, rather than by DNMT3A or DNMT3B [26,55,56].
At the molecular level, the role of UHRF1 can be summarized in the targeting of DNMT1 to replication forks by serving as a guide for DNMT1 at hemi-methylated CpG sites [25,57,58,59,60,61,62,63,64]. At the cellular level, the UHRF1/DNMT1 tandem is involved in many processes, including differentiation [65], cell senescence [66], stem cell self-renewal [67,68,69], neurogenesis [70], germinal center B-cell expansion [71], maturation of colonic T lymphocytes [72], smooth muscle plasticity [73], and induced pluripotent stem-cell reprogramming [74] and development [75,76].
1.3. UHRF1 and DNMT1, Interdependent Multi-Domain Proteins
UHRF1 is a multi-domain protein (Figure 1) including a ubiquitin-like domain (UBL), a tandem Tudor domain (TTD), a plant homeodomain (PHD), an SET- and RING-associated (SRA) domain, and a really interesting new gene (RING) domain, with the latter domain conferring the sole enzymatic activity [77]. Two-thirds of the primary sequence of UHRF1 contributes to these structural domains, suggesting that this protein fulfils multiple important roles. One of these roles is the link between the histone code and DNA methylation. Indeed, SRA is able to sense the presence of hemi-methylated DNA through its SRA domain and to recognize via the tandem Tudor domain, probably at the same time, two or three methyl groups on lysine 9 of histone H3 (H3K9me2/3) [25,61,64,78,79,80,81,82]. Accordingly, it was shown that UHRF1 can target DNMT1 for DNA methylation maintenance via binding to H3K9me2/3 or hemi-methylated CpG [83], thus ensuring a kind of security for the faithful inheritance of DNA methylation and, probably reciprocally, histone H3 methylation profiles. The PHD domain is involved in binding to unmodified H3R2 [84,85,86,87] and, thus, contributes to the binding of UHRF1 to chromatin [88]. The role of the UBL might allow UHRF1 to follow the proteasome pathway [89] and/or be involved in the interaction with the E2 conjugate to promote E3 ligase activity [90,91].
Figure 1.
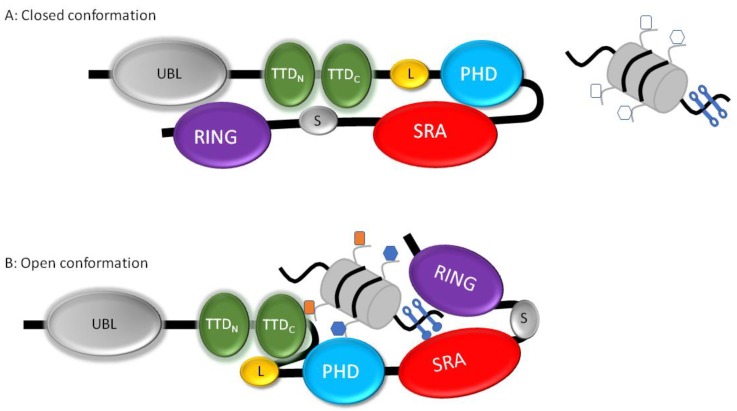
Schematic representation of ubiquitin-like containing plant homeodomain (PHD) and really interesting new gene (RING) finger domains 1 (UHRF1)’s domains with open and closed conformations. (a) Closed conformation of UHRF1. In the absence of methylated DNA (open lollipops), UHRF1 adopts a closed conformation with the spacer (S) facing the tandem Tudor domain (TTD) and the RING domain facing the ubiquitin-like domain (UBL). This interaction may represent a kind of auto-inhibitory mechanism as UBL exhibits analogy with ubiquitin. The SET- and RING-associated (SRA) domain faces the PHD domain. The linker (L) is the sequence between the TTD and PHD. (b) Open conformation of UHRF1. In the presence of hemi-methylated DNA (full lollipops on one strand) and in the presence of di/trimethylated lysine 9 on histone H3 (ocher rectangle on nucleosome) and of unmodified arginine 2 on histone H3 (blue hexagon on nucleosome), UHRF1 adopts an open conformation allowing each domain to fulfill its respective role. For instance, the RING domain can ubiquitinate histone H3 and the TTD with methylated histone H3.
DNMT1 is a large protein of 1616 amino acids (aa) [31,92,93,94,95] composed of a large regulatory N-terminal region (1000 aa) and a catalytic C-terminal region including 10 catalytic motifs that are important for the interaction with S-adenosyl methionine, the methyl group donor. The C-terminal and N-terminal regions are linked by six Lys/Gly (KG) dipeptide repeats. The N-terminal region is composed of (i) a domain able to interact with a large panel of proteins, (ii) a replication focus targeting sequence (RFTS) domain, which is a matter of very active research over the last few years, involved in the recruitment of DNMT1 to the DNA replication fork, (iii) a zinc-binding domain, (iv) two bromo-adjacent homology (BAH) domains, and (v) a nuclear localization signal (aa 191–211). The crystal structure revealed that the RFTS domain directly associates with the catalytic domain, thereby exhibiting auto-inhibition ability [96].
2. The UHRF1/DNMT1 Dialogue on Chromatin
2.1. The Ubiquitination of Histone H3 by UHRF1 as a Chromatin Anchorage for DNMT1: Model A
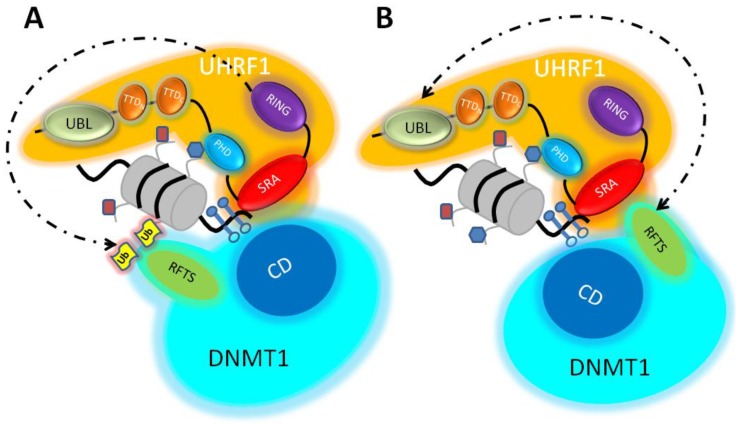
It was previously shown by several groups [96,97,98,99] that UHRF1 adopts a closed conformation with a C-terminal part (spacer aa 586–674) facing the TTD, and the SRA domain facing the PHD domain (Figure 1a). This spacer region contains a polybasic region (PBR) in the C-terminus, which prevents the recognition of H3K9me3 by the TTD domain. Therefore, in this conformation, it appears that UHRF1 is unable to bind to H3K9me2/3 via its TTD, to the unmodified R2 of histone H3 via the PHD domain, or to hemi-methylated DNA via its SRA domain [97]. However, interestingly, the presence of hemi-methylated DNA impairs the intramolecular interactions, and each domain can exert its ability to interact with its selective target (Figure 1b). It was further suggested that the docking of TTD to H3K9me3 may induce the ubiquitination activity of the RING domain toward lysines 14, 18, and 23 (K14, K18, K23) of histone H3 [97], thus giving rise to a signal for DNMT1 recruitment [100,101,102,103]. Fine-tuned regulation of H3 ubiquitination suggests that the RING finger of UHRF1 induces mono-ubiquitination of histone H3 at K14, K18, and K23 but not polyubiquitination [88,102] in spite of its ability to catalyze it [104,105]. Apparently, two mono-ubiquitinated lysines are enough for DNMT1 to bind histone H3, i.e., H3K14/K18, H3K14/K18, or H3K18/K23, with the help of the N-terminal tail of histone H3 to confer high affinity [102]. It is the RFTS domain of DNMT1 that is involved in this anchorage with similar affinities for each double mono-ubiquitinated motif [102]. The mode of ubiquitin binding is, however, different from other reported recognition patterns [106,107]. Regarding the high affinity of DNMT1 for ubiquitinated histone H3, it was suggested that this interaction gives robustness to the inheritance of DNA methylation patterns [106]. It was further proposed that these lysines could be crucial for gene expression regulation as they can be ubiquitinated (repressive effect) or acetylated by p300/CBP or GCN5 (General Control Of Amino Acid Synthesis Protein 5) which are enzymes involved in gene transcription [108].
In accordance with a role of histone H3 ubiquination in DNA methylation maintenance, depletion of ubiquitin-specific peptidase 7 (USP7) in HeLa cells enhanced histone H3 ubiquitination and led to enhancement of DNMT1 nuclear foci during DNA replication [109]. However, it was shown that USP7 stimulates both the maintenance and de novo DNA methylation activity of DNMT1 in vitro [88,110], further supporting a complex role of USP7 in DNA methylation maintenance that remains to be fully deciphered.
2.2. Domain–Domain Interactions between DNMT1 and UHRF1: Model B
There were numerous reports [24,25,60,111,112,113] showing a physical interaction between UHRF1 and DNMT1, leaving little doubt that UHRF1 communicates directly with DNMT1. Initially, two independent groups reported the interaction between UHRF1 and DNMT1 [24,60]. Domain mapping nevertheless showed some discrepancies. Indeed, we observed that the human SRA domain of UHRF1 is responsible for the interaction with human DNMT1 [60] (Figure 2b). The other group found that it is rather the PHD domain of mouse UHRF1 that is involved in DNMT1 interaction [24]. Additionally, it was recently reported that the UBL domain of UHRF1 is able to bind to the RFTS domain of DNMT1 (N-terminal region), as well with the amino-acid sequence encompassing aa 621–1616 [91,106]. The latter interaction is involved in the stimulation of the enzymatic activity of DNMT1, whereas the first (RFTS) is rather implicated in the alleviation of the auto-inhibition exerted by DNMT1 [91,106]. Three regions of DNMT1 were shown to bind to UHRF1, namely amino-acid residues 1–446, 1081–1408 in mouse [24], and 401–615 in human [60]. We previously suggested that the discrepancies might come from the species-specific difference [59]; however, it was later shown that the interacting domain in mouse DNMT1 was a region encompassing aa 291–601 corresponding to the RFTS domain [114]. In accordance, it was shown that the interacting motif of DNMT1 with UHRF1 is a region spanning from aa 380–399 [88]. Interestingly and consistently, the spacer of UHRF1, located beside the SRA domain, finds an important role as it facilitates the interaction of the SRA domain with hemi-methylated DNA and the interaction with DNMT1 [97]. It should be mentioned here that the spacer might explain the different functions of UHRF1 and UHRF2, as the latter lacks this sequence, as well as DNA methylation maintenance capacity [111,115,116]. Therefore, the most likely sites of interaction are the SRA domain and the UBL of UHRF1, with the RFTS domain of DNMT1 (Figure 2). This multiple site connection might establish a kind of dialogue between these two proteins depending on the methylation status of the DNA and/or the motion of the complex along the DNA. In accordance with this, it was shown that full-length UHRF1, the SRA domain [117], or the UBL [106] increase the preference or catalytic activity of DNMT1 for hemi-methylated DNA. This effect is dependent on the RFTS domain of DNMT1. Indeed, the RFTS of DNMT1 exerts inhibitory action on its catalytic domain, by occupying the DNA binding pocket, as shown by crystallography [112,113]. This could be evidenced by incubating DNMT1 with UHRF1 or the SRA domain prior to the addition of DNA, as UHRF1 competes with DNMT1 for binding to DNA. When DNA is added first, UHRF1 binds to DNA and does not reverse the clamped conformation of DNMT1. A model in which UHRF1 binds to hemi-methylated CpG site via its SRA domain with subsequent recruitment of DNMT1 was suggested [117]. This model supposes that UHRF1, through the SRA domain, binds firstly to DNA, which is in accordance with other studies proposing that the SRA domain moves along DNA to seek for the presence of hemi-methylated DNA [57,58]. In accordance with a histone-independent recruitment of DNMT1, it was intriguingly proposed that auto-ubiquitination of UHRF1 might serve as a docking site for DNMT1 [118].
Figure 2.
Dialogue model between UHRF1 and DNA methyltransferase 1 (DNMT1) in the presence of hemi-methylated DNA. (a) Model A: targeting of DNMT1 to chromatin via histone H3-dependent ubiquitination. Two molecules of ubiquitin (Ub) on the nucleosome, mediated by the RING domain of UHRF1 (dotted line), serve as an anchorage for DNMT1 via the replication focus targeting sequence (RFTS) domain. This interaction allows the alleviation of the auto-inhibitory activity of RFTS on the catalytic domain (CD) of DNMT1. Concomitantly, the TTD binds di/trimethylated lysine 9 on histone H3 (ocher rectangle on nucleosome) and the PDH binds unmodified arginine 2 on histone H3 (blue hexagon on nucleosome). (b) Model B: targeting of DNMT1 to chromatin via UHRF1 domain interactions. The UBL (dotted line) and SRA domain interact with the RFTS domain of UHRF1, allowing the release of the catalytic domain (CD) of DNMT1. As with model A, the TTD binds di/trimethylated lysine 9 on histone H3 (ocher rectangle on nucleosome) and the PDH binds unmodified arginine 2 on histone H3 (blue hexagon on nucleosome).
2.3. A Conciliated Model of How the UHRF1/DNMT1 Tandem Works
In light of all the studies, the following question remains: what is the correct model for the UHRF1-dependent CpG targeting by DNMT1? The majority of these studies were performed on purified isolated proteins, thus questioning the relevance of the in vivo occurring. However, if both models are right, we have to find an explanation to render them compatible.
We, therefore, propose a more complete model in which UHRF1 does not recruit DNMT1, but in which they are still together in the same large macro-molecular complex; thus, DNMT1 is in permanent presence, in close vicinity of UHRF1, as long as this complex progresses at the DNA replication fork, although a direct transfer of a CpG site from the SRA to DNMT1 appears not applicable [117]. When the SRA domain meets a hemi-methylated CpG dinucleotide, UHRF1 adopts an open conformation allowing ubiquitination of histone H3 and/or auto-ubiquitination via the RING, facilitated by the cooperation of the UBL and E2 Ube2D (ubiquitin-conjugating enzyme E2D). The ubiquitination of histone H3 and/or UHRF1 auto-ubiquitination represent an anchorage signal for the RFTS domain of DNMT1, liberating the catalytic activity of DNMT1. Meanwhile the UHRF1/TTD tandem binds to the methylated histone H3 at lysine 9. This first step is preparing DNMT1 to enter in action, i.e., to methylate the new DNA strand; however, this is not sufficient to localize it. We further propose that UHRF1, when flipping the mC through the SRA domain may undergo an allosteric change that allows creating contact zones with DNMT1 inside the large complex they belong to. This interaction facilitates accession of the catalytic center of DNMT1 to its substrate, i.e., to hemi-methylated DNA, along with the release to mC by the SRA domain, considering that steric hindrance forbids that both mCs on each DNA strand can be flipped at the same time by UHRF1 and DNMT1 [119]. We do not exclude that, when UHRF1 releases the mC, it adopts a conformational change which promotes the seeking of other hemi-methylated CpG sites, leaving behind DNMT1 to methylate the newly synthesized DNA strand. We do not exclude either that the RFTS domain, chronologically, may firstly interact with ubiquitinated histone H3 and, secondly, with the SRA domain in order to remove UHRF1 from the CpG dinucleotide, considering that DNMT1 cannot methylate the opposite DNA strand as long as UHRF1 is present on the same targeted hemi-methylated CpG.
Our model supports a repetitive dialogue between DNMT1 and UHRF1 during the synthesis (S) phase at CpG sites, and is in opposition to a model in which DNMT1 leaves the chromatin as soon as it catalyzes one enzymatic reaction, i.e., a hydrogen atom replaced by a methyl group on a cytosine. This model is particularly likely when CpG islands (repeated methylated CpG) are methylated. An interesting question arises from this model: what happens between two methylated CpG dinucleotides or between two methylated CpG islands? In our opinion, it is unlikely that DNMT1 dissociates from UHRF1 or the UHRF1 complex after each hemi-methylated CpG dinucleotide. Otherwise, it would take too much time for UHRF1 to wait for recruiting (i.e., to find) a molecule of DNMT1; furthermore, what would be the signal telling DNMT1 to join the UHRF1 complex? All that would hinder the epigenetic code replication machinery, in which UHRF1 plays the role of conductor, to follow the progression of the DNA replication fork as fast as required. In contrast, a more plausible behavior of DNMT1 and UHRF1 would be that they are in close contact in the complex while moving along the DNA, and when UHRF1, through the SRA domain, flips out the methylated cytosine, it probably communicates with DNMT1 to prepare it for recognition of the unmethylated cytosine on the opposite DNA strand. Due to allosteric constraints, UHRF1 needs to further progress to allow DNMT1 to exert its catalytic activity. Accordingly, it was proposed that the removal of the RFTS from the enzymatic pocket of DNMT1, by UHRF1, is involved in its progressive methylation property [114]. When the DNMT1/UHRF1 complex progresses along the DNA, without meeting hemi-methylated DNA, the SRA domain may not interact with the RFTS domain of DNMT1 and, in this way, DNMT1 has no enzymatic activity, avoiding undesired or unfaithful DNA methylation. However, UHRF1 may still interact with DNMT1, but through other domains previously identified [24], and this state would be a kind of standby. An interesting hypothesis would be that this standby also occurs during development, during which a faithful inheritance of DNA methylation patterns would be a brake for cell differentiation or would be an undesired event [67].
3. Concluding Remarks
In conclusion, we propose that both models are correct and compatible, but the events they are based on may be separated in time, with model A most probably happening first and model B happening just after. The following chronological model of the dialogue between UHRF1 and DNMT1 to ensure faithful inheritance of DNA methylation patterns is, therefore, proposed: hemi-methylated DNA–SRA interaction; binding of the TTD to H3K9me3; spacer facilitation of ubiquitination by the RING domain of a dual combination of lysines 14,18, and 23 of histone H3, and probably also UHRF1 auto-ubiquitination; recruitment (or preferred term interaction) of DNMT1 with mono-ubiquitinated histone H3 and/or ubiquitinated UHRF1; interaction between the SRA domain and/or UBL domain with DNMT1 (via the RFTS domain) and alleviation of the auto-inhibition of DNMT1; and, finally, methylation of the newly synthetized DNA by DNMT1 (Figure 2a,b). This model, however, is based on experimental data obtained with purified proteins, which is a situation that never occurs in vivo as evidenced by numerous studies, reporting that UHRF1 belongs to a macro-molecular complex containing a high number of partners [25,105,120]. This large macro-molecular complex is closely located to the replication fork as it associates with PCNA [18,19,83,121]. We suggest that the UHRF1/DNMT1 tandem slides along the newly synthesized DNA, immediately following the DNA replication machinery in a perpetual complex interacting mode, governed by the status of hemi-methylation of the DNA. The two models of the inheritance of DNA methylation patterns are both likely to ensure faithful DNA methylation duplication. We do not exclude that the two models can act as a kind of double check or double lock to secure fidelity of transmission of methylated profiles. Technological challenges are present that must be overcome to unravel the complexity of the dialogue (direct and/or indirect communication) between UHRF1 and DNMT1. Investigations of in situ domain–domain interactions between the two proteins will help understand when and how each model occurs. This is the price to pay to progress in the understanding of the transmission of this important epigenetic mark.
Author Contributions
C.B., M.A., A.H. and M.M. equally contributed to the writing of this review. C.B. and M.M. designed the figures.
Funding
This work was supported by institutional funds from CNRS, INSERM, Université de Strasbourg (UDS), the Labex INRT, and by grants from, INCA (INCa_4496), INCA (INCa_4454), ANR (EpivarZ, contract n° ANR-16-CE12-0013, La Ligue Nationale contre le Cancer Equipe labellisée (A.H.). Our work was supported by the Agence National de la Recherche (ANR Fluometadn), the Fondation pour la Recherche Médicale (FRM DCM20111223038), Ligue contre le Cancer and by the grant ANR-10-LABX-0030-INRT, and a French State fund managed by the Agence Nationale de la Recherche under the frame program Investissements d’Avenir ANR-10-IDEX-0002-02.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 2.Schubeler D. Function and information content of DNA methylation. Nature. 2015;517:321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- 3.Jang H.S., Shin W.J., Lee J.E., Do J.T. Cpg and non-cpg methylation in epigenetic gene regulation and brain function. Genes. 2017:8. doi: 10.3390/genes8060148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allis C.D., Jenuwein T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016;17:487–500. doi: 10.1038/nrg.2016.59. [DOI] [PubMed] [Google Scholar]
- 5.Portela A., Esteller M. Epigenetic modifications and human disease. Nat. Biotechnol. 2010;28:1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 6.Biswas S., Rao C.M. Epigenetics in cancer: Fundamentals and beyond. Pharmacol. Ther. 2017;173:118–134. doi: 10.1016/j.pharmthera.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 7.Edwards J.R., Yarychkivska O., Boulard M., Bestor T.H. DNA methylation and DNA methyltransferases. Epigenetics Chromatin. 2017;10:23. doi: 10.1186/s13072-017-0130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aran D., Toperoff G., Rosenberg M., Hellman A. Replication timing-related and gene body-specific methylation of active human genes. Hum. Mol. Genet. 2011;20:670–680. doi: 10.1093/hmg/ddq513. [DOI] [PubMed] [Google Scholar]
- 9.Aran D., Sabato S., Hellman A. DNA methylation of distal regulatory sites characterizes dysregulation of cancer genes. Genome Biol. 2013;14:R21. doi: 10.1186/gb-2013-14-3-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang X., Han H., De Carvalho D.D., Lay F.D., Jones P.A., Liang G. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014;26:577–590. doi: 10.1016/j.ccr.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jurkowska R.Z., Ceccaldi A., Zhang Y., Arimondo P.B., Jeltsch A. DNA methyltransferase assays. Methods Mol. Biol. 2011;791:157–177. doi: 10.1007/978-1-61779-316-5_13. [DOI] [PubMed] [Google Scholar]
- 12.Merlo A., Herman J.G., Mao L., Lee D.J., Gabrielson E., Burger P.C., Baylin S.B., Sidransky D. 5′ cpg island methylation is associated with transcriptional silencing of the tumour suppressor p16/cdkn2/mts1 in human cancers. Nat. Med. 1995;1:686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- 13.Pei J.H., Luo S.Q., Zhong Y., Chen J.H., Xiao H.W., Hu W.X. The association between non-hodgkin lymphoma and methylation of p73. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2011;32:1133–1138. doi: 10.1007/s13277-011-0215-0. [DOI] [PubMed] [Google Scholar]
- 14.Stefansson O.A., Jonasson J.G., Olafsdottir K., Hilmarsdottir H., Olafsdottir G., Esteller M., Johannsson O.T., Eyfjord J.E. CpG island hypermethylation of BRCA1 and loss of prb as co-occurring events in basal/triple-negative breast cancer. Epigenetics. 2011;6:638–649. doi: 10.4161/epi.6.5.15667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guan Z., Zhang J., Song S., Dai D. Promoter methylation and expression of TIMP3 gene in gastric cancer. Diagn. Pathol. 2013;8:110. doi: 10.1186/1746-1596-8-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodriguez J., Frigola J., Vendrell E., Risques R.A., Fraga M.F., Morales C., Moreno V., Esteller M., Capella G., Ribas M., et al. Chromosomal instability correlates with genome-wide DNA demethylation in human primary colorectal cancers. Cancer Res. 2006;66:8462–9468. doi: 10.1158/0008-5472.CAN-06-0293. [DOI] [PubMed] [Google Scholar]
- 17.Dawson M.A., Kouzarides T. Cancer epigenetics: From mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 18.Hervouet E., Lalier L., Debien E., Cheray M., Geairon A., Rogniaux H., Loussouarn D., Martin S.A., Vallette F.M., Cartron P.F. Disruption of Dnmt1/PCNA/UHRF1 interactions promotes tumorigenesis from human and mice glial cells. PLoS ONE. 2010;5:e11333. doi: 10.1371/journal.pone.0011333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pacaud R., Brocard E., Lalier L., Hervouet E., Vallette F.M., Cartron P.F. The Dnmt1/PCNA/UHRF1 disruption induces tumorigenesis characterized by similar genetic and epigenetic signatures. Sci. Rep. 2014;4:4230. doi: 10.1038/srep04230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nestor C.E., Ottaviano R., Reinhardt D., Cruickshanks H.A., Mjoseng H.K., McPherson R.C., Lentini A., Thomson J.P., Dunican D.S., Pennings S., et al. Rapid reprogramming of epigenetic and transcriptional profiles in mammalian culture systems. Genome Biol. 2015;16:11. doi: 10.1186/s13059-014-0576-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hopfner R., Mousli M., Garnier J.M., Redon R., du Manoir S., Chatton B., Ghyselinck N., Oudet P., Bronner C. Genomic structure and chromosomal mapping of the gene coding for ICBP90, a protein involved in the regulation of the topoisomerase iialpha gene expression. Gene. 2001;266:15–23. doi: 10.1016/S0378-1119(01)00371-7. [DOI] [PubMed] [Google Scholar]
- 22.Hopfner R., Mousli M., Jeltsch J.M., Voulgaris A., Lutz Y., Marin C., Bellocq J.P., Oudet P., Bronner C. ICBP90, a novel human ccaat binding protein, involved in the regulation of topoisomerase IIα expression. Cancer Res. 2000;60:121–128. [PubMed] [Google Scholar]
- 23.Fagerberg L., Hallstrom B.M., Oksvold P., Kampf C., Djureinovic D., Odeberg J., Habuka M., Tahmasebpoor S., Danielsson A., Edlund K., et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteom. MCP. 2014;13:397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bostick M., Kim J.K., Esteve P.O., Clark A., Pradhan S., Jacobsen S.E. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 25.Sharif J., Muto M., Takebayashi S., Suetake I., Iwamatsu A., Endo T.A., Shinga J., Mizutani-Koseki Y., Toyoda T., Okamura K., et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 26.Li E., Bestor T.H., Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-F. [DOI] [PubMed] [Google Scholar]
- 27.Muto M., Kanari Y., Kubo E., Takabe T., Kurihara T., Fujimori A., Tatsumi K. Targeted disruption of Np95 gene renders murine embryonic stem cells hypersensitive to DNA damaging agents and DNA replication blocks. J. Biol. Chem. 2002;277:34549–34555. doi: 10.1074/jbc.M205189200. [DOI] [PubMed] [Google Scholar]
- 28.Hermann A., Gowher H., Jeltsch A. Biochemistry and biology of mammalian DNA methyltransferases. Cell. Mol. Life Sci. CMLS. 2004;61:2571–2587. doi: 10.1007/s00018-004-4201-1. [DOI] [PubMed] [Google Scholar]
- 29.Alhosin M., Sharif T., Mousli M., Etienne-Selloum N., Fuhrmann G., Schini-Kerth V.B., Bronner C. Down-regulation of UHRF1, associated with re-expression of tumor suppressor genes, is a common feature of natural compounds exhibiting anti-cancer properties. J. Exp. Clin. Cancer Res. 2011;30:41. doi: 10.1186/1756-9966-30-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mohan K.N., Chaillet J.R. Cell and molecular biology of DNA methyltransferase 1. Int. Rev. Cell Mol. Biol. 2013;306:1–42. doi: 10.1016/B978-0-12-407694-5.00001-8. [DOI] [PubMed] [Google Scholar]
- 31.Jurkowska R.Z., Jeltsch A. Enzymology of mammalian DNA methyltransferases. Adv. Exp. Med. Biol. 2016;945:87–122. doi: 10.1007/978-3-319-43624-1_5. [DOI] [PubMed] [Google Scholar]
- 32.Choudhry H., Zamzami M.A., Omran Z., Wu W., Mousli M., Bronner C., Alhosin M. Targeting microRNA/UHRF1 pathways as a novel strategy for cancer therapy. Oncol. Lett. 2018;15:3–10. doi: 10.3892/ol.2017.7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gowher H., Jeltsch A. Mammalian DNA methyltransferases: New discoveries and open questions. Biochem. Soc. Trans. 2018;46:1191–1202. doi: 10.1042/BST20170574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ashraf W., Ibrahim A., Alhosin M., Zaayter L., Ouararhni K., Papin C., Ahmad T., Hamiche A., Mely Y., Bronner C., et al. The epigenetic integrator uhrf1: On the road to become a universal biomarker for cancer. Oncotarget. 2017;8:51946–51962. doi: 10.18632/oncotarget.17393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alhosin M., Omran Z., Zamzami M.A., Al-Malki A.L., Choudhry H., Mousli M., Bronner C. Signalling pathways in UHRF1-dependent regulation of tumor suppressor genes in cancer. J. Exp. Clin. Cancer Res. 2016;35:174. doi: 10.1186/s13046-016-0453-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bronner C., Krifa M., Mousli M. Increasing role of UHRF1 in the reading and inheritance of the epigenetic code as well as in tumorogenesis. Biochem Pharm. 2013;86:1643–1649. doi: 10.1016/j.bcp.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 37.Bronner C. Control of dnmt1 abundance in epigenetic inheritance by acetylation, ubiquitylation, and the histone code. Sci. Signal. 2011;4:pe3. doi: 10.1126/scisignal.2001764. [DOI] [PubMed] [Google Scholar]
- 38.Sidhu H., Capalash N. Uhrf1: The key regulator of epigenetics and molecular target for cancer therapeutics. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2017;39:1010428317692205. doi: 10.1177/1010428317692205. [DOI] [PubMed] [Google Scholar]
- 39.Beck A., Trippel F., Wagner A., Joppien S., Felle M., Vokuhl C., Schwarzmayr T., Strom T.M., von Schweinitz D., Langst G., et al. Overexpression of UHRF1 promotes silencing of tumor suppressor genes and predicts outcome in hepatoblastoma. Clin. Epigenetics. 2018;10:27. doi: 10.1186/s13148-018-0462-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boukhari A., Alhosin M., Bronner C., Sagini K., Truchot C., Sick E., Schini-Kerth V.B., Andre P., Mely Y., Mousli M., et al. CD47 activation-induced UHRF1 over-expression is associated with silencing of tumor suppressor gene p16INK4a in glioblastoma cells. Anticancer Res. 2015;35:149–157. [PubMed] [Google Scholar]
- 41.Krifa M., Alhosin M., Muller C.D., Gies J.P., Chekir-Ghedira L., Ghedira K., Mely Y., Bronner C., Mousli M. Limoniastrum guyonianum aqueous gall extract induces apoptosis in human cervical cancer cells involving p16INK4a re-expression related to UHRF1 and Dnmt1 down-regulation. J. Exp. Clin. Cancer Res. 2013;32:30. doi: 10.1186/1756-9966-32-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Achour M., Mousli M., Alhosin M., Ibrahim A., Peluso J., Muller C.D., Schini-Kerth V.B., Hamiche A., Dhe-Paganon S., Bronner C. Epigallocatechin-3-gallate up-regulates tumor suppressor gene expression via a reactive oxygen species-dependent down-regulation of UHRF1. Biochem. Biophys. Res. Commun. 2013;430:208–212. doi: 10.1016/j.bbrc.2012.11.087. [DOI] [PubMed] [Google Scholar]
- 43.Sharif T., Alhosin M., Auger C., Minker C., Kim J.H., Etienne-Selloum N., Bories P., Gronemeyer H., Lobstein A., Bronner C., et al. Aronia melanocarpa juice induces a redox-sensitive p73-related caspase 3-dependent apoptosis in human leukemia cells. PLoS ONE. 2012;7:e32526. doi: 10.1371/journal.pone.0032526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abusnina A., Keravis T., Yougbare I., Bronner C., Lugnier C. Anti-proliferative effect of curcumin on melanoma cells is mediated by pde1a inhibition that regulates the epigenetic integrator UHRF1. Mol. Nutr. Food Res. 2011;55:1677–1689. doi: 10.1002/mnfr.201100307. [DOI] [PubMed] [Google Scholar]
- 45.Abusnina A., Alhosin M., Keravis T., Muller C.D., Fuhrmann G., Bronner C., Lugnier C. Down-regulation of cyclic nucleotide phosphodiesterase PDE1A is the key event of p73 and UHRF1 deregulation in thymoquinone-induced acute lymphoblastic leukemia cell apoptosis. Cell. Signal. 2011;23:152–160. doi: 10.1016/j.cellsig.2010.08.015. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Y., Huang Z., Zhu Z., Zheng X., Liu J., Han Z., Ma X., Zhang Y. Upregulated UHRF1 promotes bladder cancer cell invasion by epigenetic silencing of KiSS1. PLoS ONE. 2014;9:e104252. doi: 10.1371/journal.pone.0104252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mousli M., Hopfner R., Abbady A.Q., Monte D., Jeanblanc M., Oudet P., Louis B., Bronner C. ICBP90 belongs to a new family of proteins with an expression that is deregulated in cancer cells. Br. J. Cancer. 2003;89:120–127. doi: 10.1038/sj.bjc.6601068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Patnaik D., Esteve P.O., Pradhan S. Targeting the set and ring-associated (SRA) domain of ubiquitin-like, phd and ring finger-containing 1 (UHRF1) for anti-cancer drug development. Oncotarget. 2018;9:26243–26258. doi: 10.18632/oncotarget.25425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jia Y., Li P., Fang L., Zhu H., Xu L., Cheng H., Zhang J., Li F., Feng Y., Li Y., et al. Negative regulation of Dnmt3a de novo DNA methylation by frequently overexpressed uhrf family proteins as a mechanism for widespread DNA hypomethylation in cancer. Cell Discov. 2016;2:16007. doi: 10.1038/celldisc.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chuang L.S., Ian H.I., Koh T.W., Ng H.H., Xu G., Li B.F. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science. 1997;277:1996–2000. doi: 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- 51.Leonhardt H., Page A.W., Weier H.U., Bestor T.H. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71:865–873. doi: 10.1016/0092-8674(92)90561-P. [DOI] [PubMed] [Google Scholar]
- 52.Spada F., Haemmer A., Kuch D., Rothbauer U., Schermelleh L., Kremmer E., Carell T., Langst G., Leonhardt H. Dnmt1 but not its interaction with the replication machinery is required for maintenance of DNA methylation in human cells. J. Cell Biol. 2007;176:565–571. doi: 10.1083/jcb.200610062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schermelleh L., Haemmer A., Spada F., Rosing N., Meilinger D., Rothbauer U., Cardoso M.C., Leonhardt H. Dynamics of Dnmt1 interaction with the replication machinery and its role in postreplicative maintenance of DNA methylation. Nucleic Acids Res. 2007;35:4301–4312. doi: 10.1093/nar/gkm432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mudbhary R., Hoshida Y., Chernyavskaya Y., Jacob V., Villanueva A., Fiel M.I., Chen X., Kojima K., Thung S., Bronson R.T., et al. UHRF1 overexpression drives DNA hypomethylation and hepatocellular carcinoma. Cancer Cell. 2014;25:196–209. doi: 10.1016/j.ccr.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fatemi M., Hermann A., Gowher H., Jeltsch A. Dnmt3a and dnmt1 functionally cooperate during de novo methylation of DNA. Eur. J. Biochem. 2002;269:4981–4984. doi: 10.1046/j.1432-1033.2002.03198.x. [DOI] [PubMed] [Google Scholar]
- 56.Jair K.W., Bachman K.E., Suzuki H., Ting A.H., Rhee I., Yen R.W., Baylin S.B., Schuebel K.E. De novo cpg island methylation in human cancer cells. Cancer Res. 2006;66:682–692. doi: 10.1158/0008-5472.CAN-05-1980. [DOI] [PubMed] [Google Scholar]
- 57.Kilin V., Gavvala K., Barthes N.P., Michel B.Y., Shin D., Boudier C., Mauffret O., Yashchuk V., Mousli M., Ruff M., et al. Dynamics of methylated cytosine flipping by UHRF1. J. Am. Chem. Soc. 2017;139:2520–2528. doi: 10.1021/jacs.7b00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Greiner V.J., Kovalenko L., Humbert N., Richert L., Birck C., Ruff M., Zaporozhets O.A., Dhe-Paganon S., Bronner C., Mely Y. Site-selective monitoring of the interaction of the sra domain of UHRF1 with target DNA sequences labeled with 2-aminopurine. Biochemistry. 2015;54:6012–6020. doi: 10.1021/acs.biochem.5b00419. [DOI] [PubMed] [Google Scholar]
- 59.Bronner C., Fuhrmann G., Chedin F.L., Macaluso M., Dhe-Paganon S. UHRF1 links the histone code and DNA methylation to ensure faithful epigenetic memory inheritance. Genet. Epigenetics. 2010;2009:29–36. doi: 10.4137/GEG.S3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Achour M., Jacq X., Ronde P., Alhosin M., Charlot C., Chataigneau T., Jeanblanc M., Macaluso M., Giordano A., Hughes A.D., et al. The interaction of the sra domain of ICBP90 with a novel domain of DNMT1 is involved in the regulation of VEGF gene expression. Oncogene. 2008;27:2187–2197. doi: 10.1038/sj.onc.1210855. [DOI] [PubMed] [Google Scholar]
- 61.Avvakumov G.V., Walker J.R., Xue S., Li Y., Duan S., Bronner C., Arrowsmith C.H., Dhe-Paganon S. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human uhrf1. Nature. 2008;455:822–825. doi: 10.1038/nature07273. [DOI] [PubMed] [Google Scholar]
- 62.Ren R., Horton J.R., Zhang X., Blumenthal R.M., Cheng X. Detecting and interpreting DNA methylation marks. Curr. Opin. Struct. Biol. 2018;53:88–99. doi: 10.1016/j.sbi.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tauber M., Fischle W. Conserved linker regions and their regulation determine multiple chromatin-binding modes of UHRF1. Nucleus. 2015;6:123–132. doi: 10.1080/19491034.2015.1026022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hashimoto H., Horton J.R., Zhang X., Bostick M., Jacobsen S.E., Cheng X. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature. 2008;455:826–829. doi: 10.1038/nature07280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smets M., Link S., Wolf P., Schneider K., Solis V., Ryan J., Meilinger D., Qin W., Leonhardt H. DNMT1 mutations found in HSANIE patients affect interaction with UHRF1 and neuronal differentiation. Hum. Mol. Genet. 2017;26:1522–1534. doi: 10.1093/hmg/ddx057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jung H.J., Byun H.O., Jee B.A., Min S., Jeoun U.W., Lee Y.K., Seo Y., Woo H.G., Yoon G. The ubiquitin-like with phd and ring finger domains 1 (UHRF1)/DNA methyltransferase 1 (DNMT1) axis is a primary regulator of cell senescence. J. Biol. Chem. 2017;292:3729–3739. doi: 10.1074/jbc.M116.750539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sen G.L., Reuter J.A., Webster D.E., Zhu L., Khavari P.A. DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature. 2010;463:563–567. doi: 10.1038/nature08683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Blanchart A., Navis A.C., Assaife-Lopes N., Usoskin D., Aranda S., Sontheimer J., Ernfors P. UHRF1 licensed self-renewal of active adult neural stem cells. Stem Cells. 2018 doi: 10.1002/stem.2889. [DOI] [PubMed] [Google Scholar]
- 69.Zhao J., Chen X., Song G., Zhang J., Liu H., Liu X. Uhrf1 controls the self-renewal versus differentiation of hematopoietic stem cells by epigenetically regulating the cell-division modes. Proc. Natl. Acad. Sci. USA. 2017;114:E142–E151. doi: 10.1073/pnas.1612967114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Murao N., Matsubara S., Matsuda T., Noguchi H., Mutoh T., Mutoh M., Koseki H., Namihira M., Nakashima K. Np95/UHRF1 regulates tumor suppressor gene expression of neural stem/precursor cells, contributing to neurogenesis in the adult mouse brain. Neurosci. Res. 2018 doi: 10.1016/j.neures.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 71.Chen C., Zhai S., Zhang L., Chen J., Long X., Qin J., Li J., Huo R., Wang X. Uhrf1 regulates germinal center B cell expansion and affinity maturation to control viral infection. J. Exp. Med. 2018;215:1437–1448. doi: 10.1084/jem.20171815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Obata Y., Furusawa Y., Endo T.A., Sharif J., Takahashi D., Atarashi K., Nakayama M., Onawa S., Fujimura Y., Takahashi M., et al. The epigenetic regulator uhrf1 facilitates the proliferation and maturation of colonic regulatory t cells. Nat. Immunol. 2014;15:571–579. doi: 10.1038/ni.2886. [DOI] [PubMed] [Google Scholar]
- 73.Elia L., Kunderfranco P., Carullo P., Vacchiano M., Farina F.M., Hall I.F., Mantero S., Panico C., Papait R., Condorelli G., et al. UHRF1 epigenetically orchestrates smooth muscle cell plasticity in arterial disease. J. Clin. Investig. 2018;128:2473–2486. doi: 10.1172/JCI96121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Milagre I., Stubbs T.M., King M.R., Spindel J., Santos F., Krueger F., Bachman M., Segonds-Pichon A., Balasubramanian S., Andrews S.R., et al. Gender differences in global but not targeted demethylation in IPSC reprogramming. Cell Rep. 2017;18:1079–1089. doi: 10.1016/j.celrep.2017.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yamashita M., Inoue K., Saeki N., Ideta-Otsuka M., Yanagihara Y., Sawada Y., Sakakibara I., Lee J., Ichikawa K., Kamei Y., et al. UHRF1 is indispensable for normal limb growth by regulating chondrocyte differentiation through specific gene expression. Development. 2018;145:dev157412. doi: 10.1242/dev.157412. [DOI] [PubMed] [Google Scholar]
- 76.Tittle R.K., Sze R., Ng A., Nuckels R.J., Swartz M.E., Anderson R.M., Bosch J., Stainier D.Y., Eberhart J.K., Gross J.M. UHRF1 and DNMT1 are required for development and maintenance of the zebrafish lens. Dev. Biol. 2011;350:50–63. doi: 10.1016/j.ydbio.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lydon-Rochelle M.T., Cardenas V., Nelson J.L., Tomashek K.M., Mueller B.A., Easterling T.R. Validity of maternal and perinatal risk factors reported on fetal death certificates. Am. J. Public Health. 2005;95:1948–1951. doi: 10.2105/AJPH.2004.044305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nady N., Lemak A., Walker J.R., Avvakumov G.V., Kareta M.S., Achour M., Xue S., Duan S., Allali-Hassani A., Zuo X., et al. Recognition of multivalent histone states associated with heterochromatin by UHRF1 protein. J. Biol. Chem. 2011;286:24300–24311. doi: 10.1074/jbc.M111.234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xie S., Jakoncic J., Qian C. UHRF1 double tudor domain and the adjacent phd finger act together to recognize k9me3-containing histone H3 tail. J. Mol. Biol. 2012;415:318–328. doi: 10.1016/j.jmb.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 80.Du J., Johnson L.M., Jacobsen S.E., Patel D.J. DNA methylation pathways and their crosstalk with histone methylation. Nat. Rev. Mol. Cell Biol. 2015;16:519–532. doi: 10.1038/nrm4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rothbart S.B., Krajewski K., Nady N., Tempel W., Xue S., Badeaux A.I., Barsyte-Lovejoy D., Martinez J.Y., Bedford M.T., Fuchs S.M., et al. Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat. Struct. Mol. Biol. 2012;19:1155–1160. doi: 10.1038/nsmb.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rothbart S.B., Dickson B.M., Ong M.S., Krajewski K., Houliston S., Kireev D.B., Arrowsmith C.H., Strahl B.D. Multivalent histone engagement by the linked tandem tudor and phd domains of UHRF1 is required for the epigenetic inheritance of DNA methylation. Genes Dev. 2013;27:1288–1298. doi: 10.1101/gad.220467.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu X., Gao Q., Li P., Zhao Q., Zhang J., Li J., Koseki H., Wong J. UHRF1 targets DNMT1 for DNA methylation through cooperative binding of hemi-methylated DNA and methylated h3k9. Nat. Commun. 2013;4:1563. doi: 10.1038/ncomms2562. [DOI] [PubMed] [Google Scholar]
- 84.Rajakumara E., Wang Z., Ma H., Hu L., Chen H., Lin Y., Guo R., Wu F., Li H., Lan F., et al. PHD finger recognition of unmodified histone H3R2 links UHRF1 to regulation of euchromatic gene expression. Mol. Cell. 2011;43:275–284. doi: 10.1016/j.molcel.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lallous N., Legrand P., McEwen A.G., Ramon-Maiques S., Samama J.P., Birck C. The PHD finger of human UHRF1 reveals a new subgroup of unmethylated histone H3 tail readers. PLoS ONE. 2011;6:e27599. doi: 10.1371/journal.pone.0027599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hu L., Li Z., Wang P., Lin Y., Xu Y. Crystal structure of PHD domain of UHRF1 and insights into recognition of unmodified histone H3 arginine residue 2. Cell Res. 2011;21:1374–1378. doi: 10.1038/cr.2011.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang C., Shen J., Yang Z., Chen P., Zhao B., Hu W., Lan W., Tong X., Wu H., Li G., et al. Structural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD finger. Cell Res. 2011;21:1379–1382. doi: 10.1038/cr.2011.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Qin W., Wolf P., Liu N., Link S., Smets M., La Mastra F., Forne I., Pichler G., Horl D., Fellinger K., et al. DNA methylation requires a DNMT1 ubiquitin interacting motif (UIM) and histone ubiquitination. Cell Res. 2015;25:911–929. doi: 10.1038/cr.2015.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ronau J.A., Beckmann J.F., Hochstrasser M. Substrate specificity of the ubiquitin and Ubl proteases. Cell Res. 2016;26:441–456. doi: 10.1038/cr.2016.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.DaRosa P.A., Harrison J.S., Zelter A., Davis T.N., Brzovic P., Kuhlman B., Klevit R.E. A bifunctional role for the UHRF1 Ubl domain in the control of hemi-methylated DNA-dependent histone ubiquitylation. Mol. Cell. 2018;72:753.e6–765.e6. doi: 10.1016/j.molcel.2018.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Foster B.M., Stolz P., Mulholland C.B., Montoya A., Kramer H., Bultmann S., Bartke T. Critical role of the UBL domain in stimulating the E3 ubiquitin ligase activity of uhrf1 toward chromatin. Mol. Cell. 2018;72:739.e9–752.e9. doi: 10.1016/j.molcel.2018.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Leonhardt H., Cardoso M.C. DNA methylation, nuclear structure, gene expression and cancer. J. Cell. Biochem. Suppl. 2000;79(Suppl. 35):78–83. doi: 10.1002/1097-4644(2000)79:35+<78::aid-jcb1129>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 93.Qin W., Leonhardt H., Pichler G. Regulation of DNA methyltransferase 1 by interactions and modifications. Nucleus. 2011;2:392–402. doi: 10.4161/nucl.2.5.17928. [DOI] [PubMed] [Google Scholar]
- 94.Jeltsch A., Jurkowska R.Z. New concepts in DNA methylation. Trends Biochem. Sci. 2014;39:310–318. doi: 10.1016/j.tibs.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 95.Jeltsch A., Jurkowska R.Z. Allosteric control of mammalian DNA methyltransferases—A new regulatory paradigm. Nucleic Acids Res. 2016;44:8556–8575. doi: 10.1093/nar/gkw723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang Z.M., Liu S., Lin K., Luo Y., Perry J.J., Wang Y., Song J. Crystal structure of human DNA methyltransferase 1. J. Mol. Biol. 2015;427:2520–2531. doi: 10.1016/j.jmb.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fang J., Cheng J., Wang J., Zhang Q., Liu M., Gong R., Wang P., Zhang X., Feng Y., Lan W., et al. Hemi-methylated DNA opens a closed conformation of uhrf1 to facilitate its histone recognition. Nat. Commun. 2016;7:11197. doi: 10.1038/ncomms11197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gelato K.A., Tauber M., Ong M.S., Winter S., Hiragami-Hamada K., Sindlinger J., Lemak A., Bultsma Y., Houliston S., Schwarzer D., et al. Accessibility of different histone H3-binding domains of UHRF1 is allosterically regulated by phosphatidylinositol 5-phosphate. Mol. Cell. 2014;54:905–919. doi: 10.1016/j.molcel.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 99.Gao L., Tan X.F., Zhang S., Wu T., Zhang Z.M., Ai H.W., Song J. An intramolecular interaction of UHRF1 reveals dual control for its histone association. Structure. 2018;26:304.e3–311.e3. doi: 10.1016/j.str.2017.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Misaki T., Yamaguchi L., Sun J., Orii M., Nishiyama A., Nakanishi M. The replication foci targeting sequence (RFTS) of DNMT1 functions as a potent histone H3 binding domain regulated by autoinhibition. Biochem. Biophys. Res. Commun. 2016;470:741–747. doi: 10.1016/j.bbrc.2016.01.029. [DOI] [PubMed] [Google Scholar]
- 101.Nishiyama A., Yamaguchi L., Sharif J., Johmura Y., Kawamura T., Nakanishi K., Shimamura S., Arita K., Kodama T., Ishikawa F., et al. Uhrf1-dependent H3K23 ubiquitylation couples maintenance DNA methylation and replication. Nature. 2013;502:249–253. doi: 10.1038/nature12488. [DOI] [PubMed] [Google Scholar]
- 102.Ishiyama S., Nishiyama A., Saeki Y., Moritsugu K., Morimoto D., Yamaguchi L., Arai N., Matsumura R., Kawakami T., Mishima Y., et al. Structure of the dnmt1 reader module complexed with a unique two-mono-ubiquitin mark on histone H3 reveals the basis for DNA methylation maintenance. Mol. Cell. 2017;68:350.e357–360.e357. doi: 10.1016/j.molcel.2017.09.037. [DOI] [PubMed] [Google Scholar]
- 103.Harrison J.S., Cornett E.M., Goldfarb D., DaRosa P.A., Li Z.M., Yan F., Dickson B.M., Guo A.H., Cantu D.V., Kaustov L., et al. Hemi-methylated DNA regulates DNA methylation inheritance through allosteric activation of H3 ubiquitylation by UHRF1. eLife. 2016;5:e17101. doi: 10.7554/eLife.17101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jenkins Y., Markovtsov V., Lang W., Sharma P., Pearsall D., Warner J., Franci C., Huang B., Huang J., Yam G.C., et al. Critical role of the ubiquitin ligase activity of UHRF1, a nuclear ring finger protein, in tumor cell growth. Mol. Biol. Cell. 2005;16:5621–5629. doi: 10.1091/mbc.e05-03-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ibrahim A., Alhosin M., Papin C., Ouararhni K., Omran Z., Zamzami M.A., Al-Malki A.L., Choudhry H., Mely Y., Hamiche A., et al. Thymoquinone challenges UHRF1 to commit auto-ubiquitination: A key event for apoptosis induction in cancer cells. Oncotarget. 2018;9:28599–28611. doi: 10.18632/oncotarget.25583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li T., Wang L., Du Y., Xie S., Yang X., Lian F., Zhou Z., Qian C. Structural and mechanistic insights into uhrf1-mediated DNMT1 activation in the maintenance DNA methylation. Nucleic Acids Res. 2018;46:3218–3231. doi: 10.1093/nar/gky104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Komander D., Rape M. The ubiquitin code. Annu. Rev. Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 108.Peterson C.L., Laniel M.A. Histones and histone modifications. Curr. Biol. 2004;14:R546–R551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 109.Yamaguchi L., Nishiyama A., Misaki T., Johmura Y., Ueda J., Arita K., Nagao K., Obuse C., Nakanishi M. Usp7-dependent histone h3 deubiquitylation regulates maintenance of DNA methylation. Sci. Rep. 2017;7:55. doi: 10.1038/s41598-017-00136-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Felle M., Joppien S., Nemeth A., Diermeier S., Thalhammer V., Dobner T., Kremmer E., Kappler R., Langst G. The usp7/dnmt1 complex stimulates the DNA methylation activity of DNMT1 and regulates the stability of UHRF1. Nucleic Acids Res. 2011;39:8355–8365. doi: 10.1093/nar/gkr528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bronner C., Achour M., Arima Y., Chataigneau T., Saya H., Schini-Kerth V.B. The UHRF family: Oncogenes that are drugable targets for cancer therapy in the near future? Pharmacol. Ther. 2007;115:419–434. doi: 10.1016/j.pharmthera.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 112.Takeshita K., Suetake I., Yamashita E., Suga M., Narita H., Nakagawa A., Tajima S. Structural insight into maintenance methylation by mouse DNA methyltransferase 1 (DNMT1) Proc. Natl. Acad. Sci. USA. 2011;108:9055–9059. doi: 10.1073/pnas.1019629108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Syeda F., Fagan R.L., Wean M., Avvakumov G.V., Walker J.R., Xue S., Dhe-Paganon S., Brenner C. The replication focus targeting sequence (RFTS) domain is a DNA-competitive inhibitor of DNMT1. J. Biol. Chem. 2011;286:15344–15351. doi: 10.1074/jbc.M110.209882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Berkyurek A.C., Suetake I., Arita K., Takeshita K., Nakagawa A., Shirakawa M., Tajima S. The DNA methyltransferase DNMT1 directly interacts with the set and ring finger-associated (SRA) domain of the multifunctional protein UHRF1 to facilitate accession of the catalytic center to hemi-methylated DNA. J. Biol. Chem. 2014;289:379–386. doi: 10.1074/jbc.M113.523209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pichler G., Wolf P., Schmidt C.S., Meilinger D., Schneider K., Frauer C., Fellinger K., Rottach A., Leonhardt H. Cooperative DNA and histone binding by UHRF2 links the two major repressive epigenetic pathways. J. Cell. Biochem. 2011;112:2585–2593. doi: 10.1002/jcb.23185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhang J., Gao Q., Li P., Liu X., Jia Y., Wu W., Li J., Dong S., Koseki H., Wong J. S phase-dependent interaction with DNMT1 dictates the role of UHRF1 but not UHRF2 in DNA methylation maintenance. Cell Res. 2011;21:1723–1739. doi: 10.1038/cr.2011.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bashtrykov P., Jankevicius G., Jurkowska R.Z., Ragozin S., Jeltsch A. The uhrf1 protein stimulates the activity and specificity of the maintenance DNA methyltransferase DNMT1 by an allosteric mechanism. J. Biol. Chem. 2014;289:4106–4115. doi: 10.1074/jbc.M113.528893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Vaughan R.M., Dickson B.M., Whelihan M.F., Johnstone A.L., Cornett E.M., Cheek M.A., Ausherman C.A., Cowles M.W., Sun Z.W., Rothbart S.B. Chromatin structure and its chemical modifications regulate the ubiquitin ligase substrate selectivity of UHRF1. Proc. Natl. Acad. Sci. USA. 2018;115:8775–8780. doi: 10.1073/pnas.1806373115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Arita K., Ariyoshi M., Tochio H., Nakamura Y., Shirakawa M. Recognition of hemi-methylated DNA by the sra protein UHRF1 by a base-flipping mechanism. Nature. 2008;455:818–821. doi: 10.1038/nature07249. [DOI] [PubMed] [Google Scholar]
- 120.Ferry L., Fournier A., Tsusaka T., Adelmant G., Shimazu T., Matano S., Kirsh O., Amouroux R., Dohmae N., Suzuki T., et al. Methylation of DNA ligase 1 by G9A/GLP recruits UHRF1 to replicating DNA and regulates DNA methylation. Mol. Cell. 2017;67:550.e5–565.e5. doi: 10.1016/j.molcel.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 121.Miura M., Watanabe H., Sasaki T., Tatsumi K., Muto M. Dynamic changes in subnuclear Np95 location during the cell cycle and its spatial relationship with DNA replication foci. Exp. Cell Res. 2001;263:202–208. doi: 10.1006/excr.2000.5115. [DOI] [PubMed] [Google Scholar]