Abstract
Epithelial cells and differentiated fiber cells represent distinct compartments in the ocular lens. While previous studies have revealed proteins that are preferentially expressed in epithelial vs. fiber cells, a comprehensive proteomics library comparing the molecular compositions of epithelial vs. fiber cells is essential for understanding lens formation, function, disease and regenerative potential, and for efficient differentiation of pluripotent stem cells for modeling of lens development and pathology in vitro. To compare protein compositions between the lens epithelium and fibers, we employed tandem mass spectrometry (2D-LC/MS) analysis of microdissected mouse P0.5 lenses. Functional classifications of the top 525 identified proteins into gene ontology categories by molecular processes and subcellular localizations, were adapted for the lens. Expression levels of both epithelial and fiber proteomes were compared with whole lens proteome and mRNA levels using E14.5, E16.5, E18.5, and P0.5 RNA-Seq data sets. During this developmental time window, multiple complex biosynthetic and catabolic processes generate the molecular and structural foundation for lens transparency. As expected, crystallins showed a high correlation between their mRNA and protein levels. Comprehensive data analysis confirmed and/or predicted roles for transcription factors (TFs), RNA-binding proteins (e.g. Carhsp1), translational apparatus including ribosomal heterogeneity and initiation factors, microtubules, cytoskeletal [e.g. non-muscle myosin IIA heavy chain (Myh9) and βB2-spectrin (Sptbn2)] and membrane proteins in lens formation and maturation. Our data highlighted many proteins with unknown function in the lens that were preferentially enriched in epithelium or fibers, setting the stage for future studies to further dissect the roles of these proteins in fiber cell differentiation vs. epithelial cell maintenance. In conclusion, the present proteomic datasets represent the first mouse lens epithelium and fiber cell proteomes, establish comparative analyses of protein and RNA-Seq data, and characterize the major proteome remodeling required to form the mature lens fiber cells.
Keywords: Differentiation, lens, mass spectrometry, RNA-Seq, transcription factors, transcriptome, proteome
1. Introduction
The ocular lens represents a unique system to probe mechanisms of cellular differentiation, cell and membrane remodeling, translational regulation, protein quality control, and protein transport (Bassnett and Sikic, 2017; Cvekl and Zhang, 2017; Horwitz, 2003; Morishita and Mizushima, 2016; Schey et al., 2014). These processes evolved in concert to support lens transparency and its refractive power (Bassnett et al., 2011). Lens proteins formed during early lens morphogenesis persist throughout the entire lens lifespan, and studies of protein aging in the lens serve as a paradigm to understand a variety of protein aggregation and misfolding diseases (Toyama and Hetzer, 2013), including age-related cataracts, defined as any opacity in the lens (Michael and Bron, 2011; Sharma and Santhoshkumar, 2009; Truscott and Friedrich, 2016).
Age-related cataracts are found in over 60% of individuals over the age of 85, and females are at much higher risk compared to males (nei.nih.gov/eyedata/cataract) (Klein et al., 1998). Cataract surgery to replace opaque lenses with artificial intraocular lenses is the number one vision-related Medicare cost in the United States. Despite the success of cataract surgeries, cataracts remain the leading cause of blindness in the world, and cataract surgery is not without risk. Potential side effects include secondary cataract formation (Petrash, 2013; Walker and Menko, 2009; West-Mays et al., 2010; Wormstone and Eldred, 2016), infection or bleeding in the eye, increased intraocular pressure, retinal detachment and droopy eyelid. Use of small molecules to prevent and/or delay the onset of human cataract formation represents an ongoing challenge for lens research (Barnes and Quinlan, 2017; Quinlan, 2015), and no non-surgical interventions are currently available. To facilitate these drug development studies, in vitro cultures from human embryonic stem cells and induced pluripotent (ES/iPS) cells may become an advantageous system to generate lens-like organoids for both high throughput drug screening and mechanistic studies. Although considerable progress has been achieved in this field in recent years (Fu et al., 2017; Murphy et al., 2018; Yang et al., 2010), in vitro generated lens-like organoids still need to be fully characterized in order to evaluate how closely culture systems mimic normal lens development and to compare the fidelity of different culture and differentiation systems.
A rapidly growing number of proteomic studies have generated data using bovine (Wang and Schey, 2009, 2015; Wilmarth et al., 2009), chicken (Chen et al., 2016; Wilmarth et al., 2009), human (Wang et al., 2017; Wang et al., 2013; Wenke et al., 2016), mouse (Bassnett et al., 2009; Khan et al., 2018; Liu et al., 2015; Whitson et al., 2017), and zebrafish (Greiling et al., 2009; Posner et al., 2017) lens materials. In addition, several mammalian cataract models were recently subjected to in depth proteomic analyses (Shang et al., 2014; Whitson et al., 2017). However, very few of these studies separately probed the epithelial and fiber cell compartments. Although there is an increasing number of lens “quantitative” RNA-Seq datasets (Audette et al., 2016; Cavalheiro et al., 2017; Hoang et al., 2014; Khan et al., 2015; Sun et al., 2015b), the attempts, so far, to analyze and connect both the lens proteomes and transcriptomes are very limited. These combined studies are required to better understand the entire transcriptional-translational cascade as the foundation for lens differentiation and remodeling to achieve and maintain lens transparency and refractive power.
To address these gaps in our understanding of the molecular composition of the mammalian lens, we conducted parallel analyses of mouse lens epithelial and lens fiber cell proteomes and transcriptomes and presented the first comprehensive comparative analysis of both lens compartments. The present data and the accompanying RNA-Seq study (Zhao et al., 2018) established multiple reference datasets for future studies of human lens proteomes and transcriptomes, including resources to aid The Human Cell Atlas project (Rozenblatt-Rosen et al., 2017). Importantly, the analysis of our data allowed the segregation of proteins needed for epithelial cell maintenance vs. fiber cell differentiation. These datasets pave the road for future studies of mammalian lens proteomes and stem-cell-derived lens-like organoids to determine whether these cultured structures undergo the correct programming to create distinct epithelial and fiber cell populations.
2. Materials and Methods
2.1. Tissue samples and protein isolation
The lens epithelium and fiber were microdissected from P0.5 mice (CD1 strain) as described in detail previously (Zhao et al., 2018). Seventy-four frozen P0.5 epithelium, 74 P0.5 cortical fiber, and 22 whole lens samples from mice were dispersed by adding 0.25, 1.0, and 1.0 ml of 100 mM ammonium bicarbonate buffer, respectively, followed by 3 × 5 sec probe sonication (60 Sonic Dismembrator, Fisher Scientific) with 30 sec of cooling on ice between treatments. The smaller volume of homogenization buffer used for the preparation of the epithelial sample was to partially adjust for the smaller amount of tissue in this sample. Samples were then further homogenized using a 1 ml Tenbroeck tissue grinder (Wheaton), and the protein concentrations were determined using bovine serum albumin as a standard (BCA assay, Thermo Scientific). Epithelium, cortical fiber, and whole lens samples yielded 0.168, 4.91, and 1.80 mg of total protein, respectively. One hundred microgram portions of each sample were then trypsinized overnight at 37°C with agitation at an enzyme:substrate ratio of 1:25 following reduction and alkylation in dithiothreitol/iodoacetamide in the presence of ProteaseMax™ detergent as recommended by the man ufacturer (ProMega). Trifluoroacetic acid was then added to a final 1% concentration, and samples were centrifuged at 14,000xg for 10 minutes. The supernatant was removed, and samples were dried by vacuum centrifugation. 50 µg portions were then dissolved in 21 µl of 10mM ammonium acetate, pH 9.0 (buffer A) in preparation for two-dimensional liquid chromatography/mass spectrometric analysis (2D-LC/MS).
2.2. 2D-LC/MS analysis
Forty-eight microgram (20 µl) portions of digests from epithelium, fibers, and whole lenses from P0.5 mice were separated by 2D-LC/MS using two rounds of reversed phase chromatography. The first dimension separation was performed at pH9 in buffer A, and the second dimension separation was performed in 0.1% formic acid using an automated nanoflow liquid chromatography method as previous described (Whitson et al., 2017), except peptides were eluted from the first dimension column using 14, 17, 20, 22, 24, 26, 28, 30, 40, and 90% acetonitrile concentrations in buffer A. The 10 second-dimension separations were performed with 100 minutes of data collection using an Orbitrap Fusion mass spectrometer (Thermo Scientific) with an extra 20 minutes for column washing and re-equilibration for a total of 20 hours analysis time for each digest. Data was collected using Orbitrap profile survey scans between 375–1500m/z at a resolution of 120,000, a user defined m/z = 445.12 polysiloxane ion lock mass, 50ms maximum injection time (MIT), and automatic gain control (AGC) = 4 ×105, and 3 seconds dwell time. Data-dependent MS2 scans were triggered using a minimum intensity of 5 × 103, MIPS filter set for peptides, charge states from +2 to +7, exclusion of isotopic peaks, and dynamic exclusion set at a repeat count of 1, duration of 60 s, and +/−10ppm mass tolerance. Peptides were isolated using the quadrupole at a 1.6m/z isolation width, fragmented by collision induced dissociation (CID) at a normalized collision energy of 35%, q activation = 0.25, MIT = 300 ms, AGC =5 ×103, and rapid scan rate in the ion trap in centroid mode.
2.3. Data Analysis
The ten raw files from each of the three 2D-LC/MS analyses were loaded into MaxQuant (version 1.6.10) for peptide identification and MS1 feature detection (Cox and Mann, 2008). The first-dimension fractions were combined for each biological sample. Default parameters were used with two exceptions: iBAQ (intensity-based absolute quantification) quantification (Krey et al., 2014; Schwanhausser et al., 2011) was turned on and matching between runs was turned off. The protein database was a canonical reference proteome for mouse (UP000000589) with 22,276 sequences downloaded from www.UniProt.org (version 2017.12) using software available at https://github.com/Delan-Huang/Reference_Proteome_Manager.git. There were 840,765 total MS2 scans acquired. Of those, 182,145 had PSMs passing the 1% false discover rate (FDR). The parsimonious number of identified proteins was 5,633 across the three samples at a 1% FDR. Protein relative abundance estimates were based on iBAQ values (Schwanhausser et al., 2011). Proteins were ranked by decreasing iBAQ values, and relative abundances were expressed as percentages of the sum of all iBAQ values within each sample. In order to categorize and interpret results, the rich annotations available for mouse Swiss-Prot (reviewed) proteins were downloaded from UniProt.org. In-house scripts parsed a variety of annotation information (GO terms, gene names, keywords, and pathways) from the flat text format files and annotations were matched to the identified proteins. The annotations of interest were added to the main MaxQuant protein Groups table to facilitate filtering proteins by annotation information and enable manual categorizations. The mass spectrometry proteomics data has been deposited to the ProteomeXchange Consortium via the PRIDE (Vizcaino et al., 2016) partner repository with the dataset identifier PXD009639.
2.4. Annotations of the top abundant proteins in lens epithelium and fibers
The top 525 abundant proteins in lens epithelium and lens fiber were classified into 26 lens functional categories, based on knowledge and literature in the field. The functional categories were further merged into 8 larger groups: nucleus and DNA, protein synthesis, membrane, cytoskeleton, signaling, mitochondria, degradation and recycling, and metabolism.
2.5. Comparisons of proteomic and RNA-Seq data from P0.5 epithelium and fibers
Only unique genes that were detected by both proteomic and RNA-Seq data were used for the analysis, including 4584 and 3977 genes in lens epithelium and fibers, respectively. The log2 (Fragments Per Kilobase of the transcript per Million mapped reads (FPKM)) of P0.5 epithelium and fibers averaged over three replicates from RNA-Seq data (Zhao et al., 2018) and the log2 (iBAQ) values from the proteomic data were used as mRNA and protein relative expression levels for each gene and used for computing the Spearman’s correlations. The Spearman correlation coefficients of mRNA and protein levels of crystallin genes were also calculated.
2.6. Transcription factors analysis
To identify transcription factors expressed uniquely in lens epithelium or fiber, 1,484 mouse TFs were downloaded from the AnimalTFDB (Zhang et al., 2015) site (http://bioinfo.life.hust.edu.cn/AnimalTFDB/). Among them, 131 TFs were detected by both RNA-Seq and proteomic data, 69 of which were supported by literature for known or potential DNA binding roles in lens and thus selected for further analysis here.
2.7. Enriched proteins in the epithelial and fiber compartments
More than 200 proteins from 6 major categories (RNA-binding, microtubules, cytoskeleton, membrane, initiation factors, ribosomes) were collected from the top 525 epithelial or fiber proteins for further rank-based analysis. Epithelium enriched proteins were identified by determining the ratios between the fiber rank and the epithelium rank (Enrichment ratios for epithelium enriched proteins). Fiber enriched proteins were revealed by the ratios between the epithelium rank and the fiber rank (Enrichment ratios for fiber enriched proteins). All proteins with Enrichment ratios over 2 were identified as epithelium-enriched or fiber-enriched proteins. All other proteins are classified as not enriched. We then calculated the percentages of epithelium, fiber, and non-enriched proteins within each category.
2.8. Immunofluorescence
Primary antibodies used for immunofluorescence are anti-CARHSP1 (Novus Biologicals, NBP1–31660, 1:100), anti-Myh9 (Abcam, ab75590, 1:100), and anti-β-spectrin II (BD Biosciences, 611451, 1:100). Whole eyeballs were collected from P0.5 CD1 mice (Charles River Laboratories). A small opening was made at the corneal–scleral junction to facilitate fixative penetration. Eyeballs were then fixed for 4 hours in 1% paraformaldehyde in 1X PBS at 4°C. After fixation, samples were washed in ice- cold 1X PBS, cryoprotected in 30% sucrose and frozen in optimal cutting temperature (OCT) medium (Sakura Finetek, Torrance, CA, USA) in the anterior-posterior orientation. Frozen tissue blocks were stored at −80°C until sectioning. Frozen sections (10 µm thick) were collected with a Leica CM1950 cryostat (Wetzlar, Germany). The tissue slides were washed in 1X PBS plus 0.1% Triton X-100 and permeabilized 1X PBS plus 0.3% Triton X-100 for 30 minutes. Tissue slides were then blocked for 60 minutes in 3% bovine serum albumin plus 1% goat serum in 1X PBS. After blocking, sections were then hybridized with primary antibody. Tissues were incubated with NMIIA/Myh9 antibody for 15 minutes at room temperature then for 45 minutes at 37°C. Tissues were incubated with β-spectrin II antibody for 15 minutes at room temperature then overnight at 4°C. For Carhsp1 antibody, tissues wer e also incubated for 15 minutes at room temperature then overnight at 4°C. Slides were then washed and incubated with anti-rabbit Alexa Fluor 488-conjugated secondary antibody (1:100 dilution) and Alexa 555-phalloidin (F-actin stain, 1:100 dilution; Life Technologies) for 2 hours at room temperature. Finally, slides were washed and mounted with a DAPI-containing mounting media (Vectashield) and1.5mm thick glass coverslips. Images for NMIIA/Myh9 and β-spectrin II were taken with confocal imaging system Leica SP8 ×10 air or x63 oil-immersion objectives. Images for CARHSP1 were taken with Carl Zeiss inverted fluorescence microscope x10 or x63 oil-immersion objectives. For x63 oil-immersion the quantification was done on stacks of 41 optical sections with Z spacing of 0.2 µm. Three-dimensional image data were acquired using the Zeiss Axio Observer CLEM microscope (Carl Zeiss) with a Zeiss AxioCam MRm Black/White camera using Axiovision Software 10X, 1.0 NA and 60X 1.4 NA objectives and filters for DAPI, FITC and Cy3. Slides were evaluated at 10X magnification to review overall section quality. All 41 image z-stacks were compressed into one image using Maximum Projection in ImageJ. Three biological replicates were used for each protein staining and representative images were shown.
3. Results and Discussion
3.1. Global analysis of newborn mouse lens epithelial and fiber cell proteomes
Our experiments revealed lists of proteins present in lens epithelium and lens fibers, including 4,951 and 4,281 unique entries (by iBAQ values (intensity-based absolute quantification; the sum of all MS1 features associated with a particular protein normalized by the predicted number of detectible tryptic peptides for that protein. iBAQ values have been shown to be well correlated with absolute protein abundances (Krey et al., 2014; Schwanhausser et al., 2011)) >0), with 3,766 in common and 1,185 epithelium and 515 fiber proteins unique to the two lens compartments (Fig. 1A). The total number of proteins identified in the epithelium and fiber samples was 5,466. For comparison, an independent proteomic analysis of the whole lens identified a total of 4,438 proteins, including 125 proteins with very low abundances (they are proteins with iBAQ values of bottom 98%−100% total proteins in the whole lens) not detected in the microdissected preparations (data not shown). All proteins detected are listed in Supplemental Table S2. Analysis of cumulative percentage showed that 54 and 525 proteins account for 90% of the total iBAQ values for lens fiber and epithelium proteomes, respectively (Fig. 1B). To reach the 95% coverage, 196 lens fiber and 969 epithelium proteins were needed, respectively. The main reason for these differences was high levels of crystallin proteins in the lens fibers (see 3.2 for details).
Fig. 1. Global analysis of P0.5 mouse lens epithelial and lens fiber proteomes.
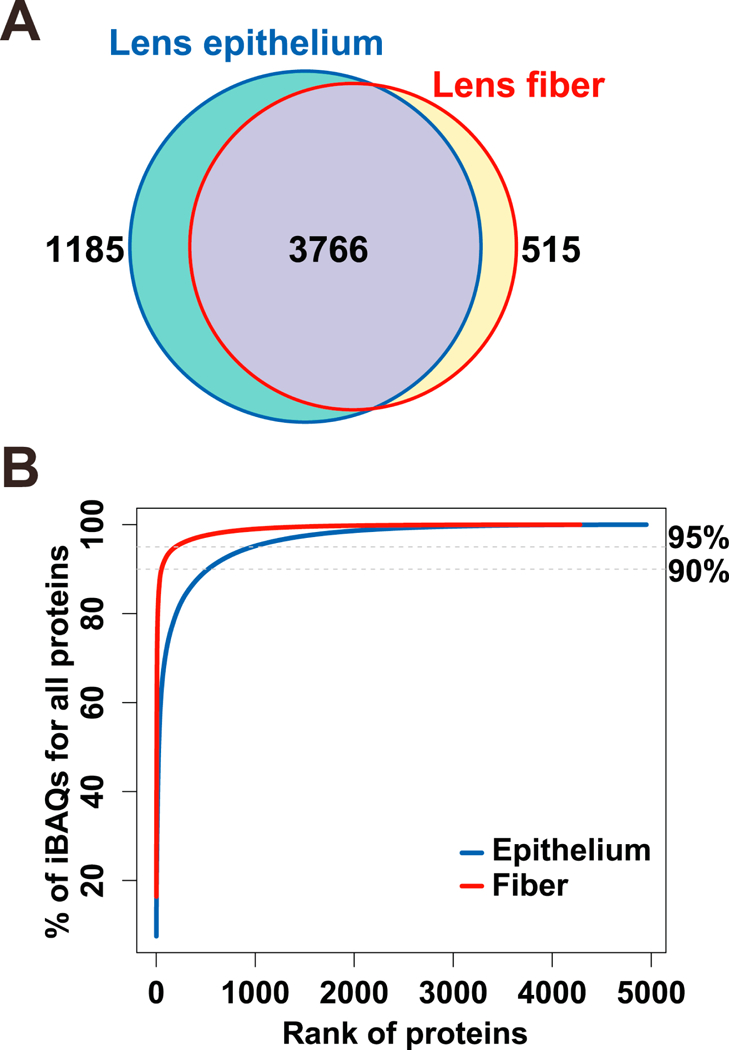
(A) Venn diagram to compare proteomes of lens epithelium and lens fibers. (B) Saturation analysis as a function of relative abundance rank of individual proteins.
To better understand which proteins contribute highly to both proteomes, we reasoned that a comparable number of proteins should be analyzed. In lens epithelium, 525 and 657 proteins, respectively, represented 90% and 92% percentiles, while 525 fiber cell proteins represented nearly 98% of all fiber cell proteins. We decided to use the top 525 proteins in each compartment. The proteins in both compartments were initially annotated using gene ontology (GOs) and then further manually curated to emphasize basic knowledge of lens structure and function. We found that lens-adjusted annotations of these proteins resulted in only 26 categories (Fig. 2, Table S1). These functional groups were subsequently combined into eight larger categories: nucleus and DNA, protein synthesis, membrane (including the extracellular compartment), microtubule, cytoskeleton, signaling, mitochondria, degradation and recycling, and metabolism (Fig. 2). The largest protein groups showed distinct proportions of epithelium- and fiber-enriched proteins (see section 2.7), including RNA-binding proteins (epithelium >> fibers), mitochondria (epithelium >> fibers), ribosomes (fibers >> epithelium), proteasomes (fibers > epithelium), membrane (fibers >> epithelium), microtubules (fibers >> epithelium) and initiation factors (eIFs, fibers >> epithelium) (shown in Table S3). Expression of globin genes and proteins in mouse lens using immunofluorescence, immunobloting, and RNA analysis was shown in detail elsewhere (Mansergh et al., 2008). It is interesting that only one protein detected in the epithelium and six proteins detected in fibers could not be annotated due to the lack of functional data.
Fig. 2. Cellular process and cell compartment analysis of top 525 most abundant proteins comprising the lens epithelial and lens fiber proteomes.
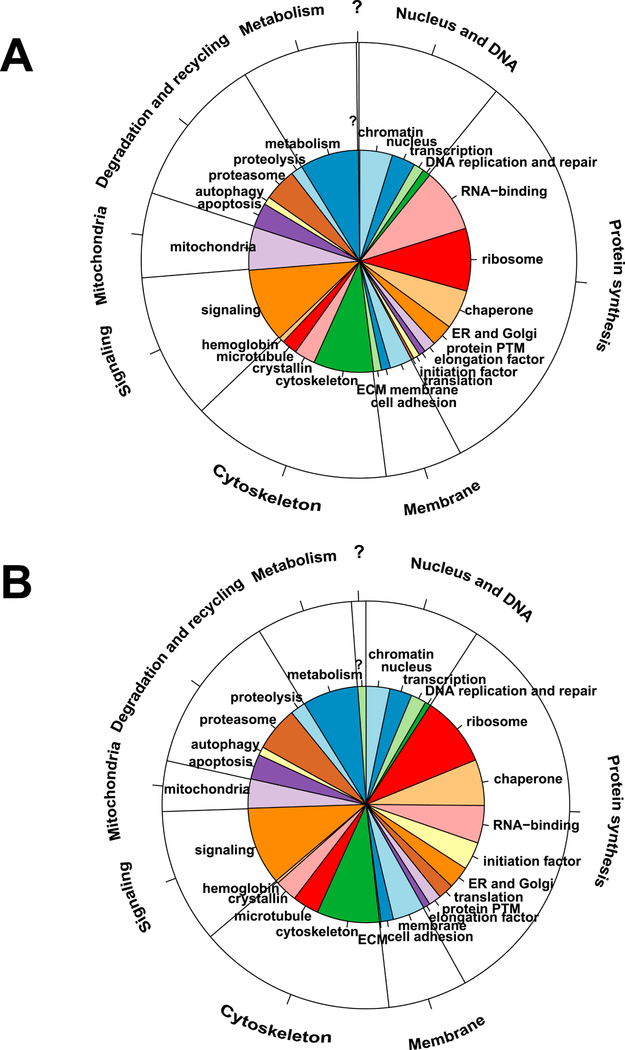
(A) Epithelium data/pie diagram. (B) Fiber cell data/pie diagram. The inner functional categories are the 26 lens terms. The annotations in the outer layer are the 8 (not including “no annotation” [?]) larger functional groups. Manually curated functional annotations of individual proteins were chosen based on known lens biology.
Use of the top 525 proteins in the detailed analysis was done to improve proteomics reproducibility since the most variable proteins in proteomics experiments are found among the higher-ranked, lower abundance proteins. To further analyze these data, we first focused on the comparison between protein and mRNA levels globally, followed by detailed analysis of selected groups, including DNA-binding transcription factors, RNA-binding proteins, components of ribosomes and translational systems, cytoskeleton, and membrane proteins.
3.2. Comparison of proteomics and RNA-Seq data
To understand the relationship between mRNA and protein levels, we calculated the Spearman’s correlation coefficients between them for both lens epithelium and fibers (Fig. 3A). The top 90% abundant proteins (n=54) in lens fibers showed a strong positive correlation to the mRNA levels (r=0.66), while in lens epithelium (n=525) the correlation was weaker (r=0.37) (Fig. 3A). Overall, a significant positive correlation between protein and mRNA levels was observed (Fig. 3A, r=0.5 and r=0.46 in fiber and epithelium including all proteins, both p values <2.2e-16). Excluding the highly abundant crystallin proteins, we found that in lens epithelium and fiber the correlations between protein and mRNA levels were r=0.46 and r=0.50, respectively with p values <2.2e-16 (Fig. 3B and Fig. 3C). The crystallin proteins (n=15) had exhibited stronger correlations in both epithelium and fiber (r=0.83 and r=0.75, respectively), as shown in Fig. 3D.
Fig. 3. Relationship of mRNA and protein abundances.
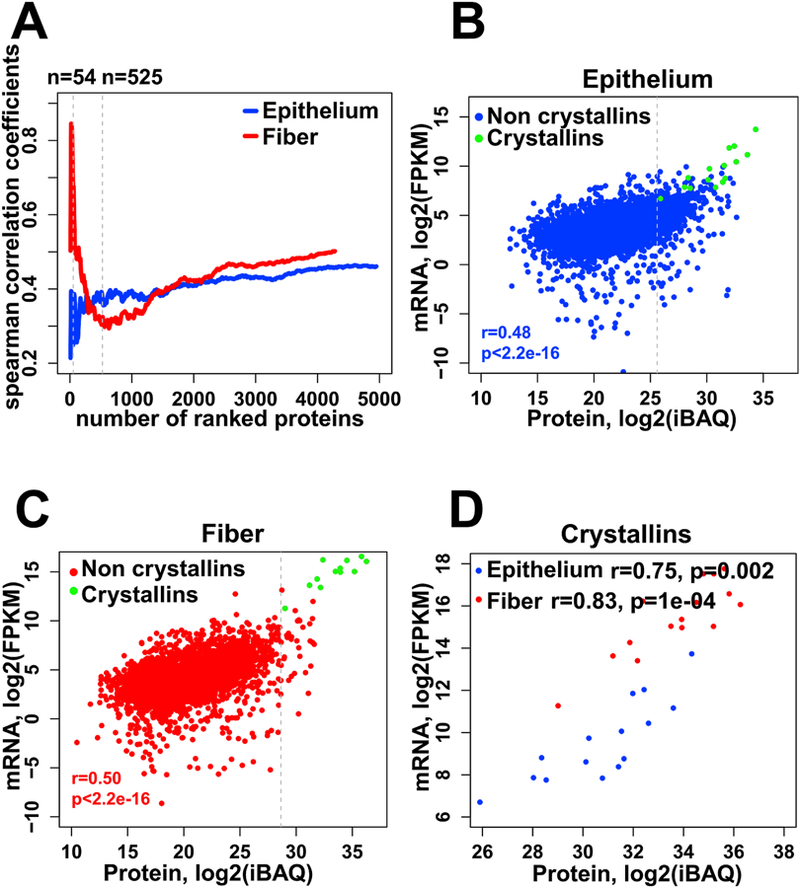
(A) A plot of the Spearman’s correlation coefficients computed for the top ranked proteins, sorted by decreasing relative abundance, for fiber (red) cells and epithelium (blue) cells. (B, C) Scatter plots of the correlation between protein and mRNA levels for epithelium (B) and lens fibers (C). Crystallins are labeled green and non crystallins are labeled blue and red for lens epithelium and fiber respectively. The Spearman’s correlation coefficients shown are for non-crystallin proteins. (D) Correlation between protein and mRNA levels of 15 crystallins in epithelium (blue) and fiber (red). The protein and mRNA levels are calculated as described in the Materials and Methods section 2.5, and plotted here in log2 transformation.
Lens crystallins represented as much as 78% of our fiber cell proteome. The top non-crystallin proteins were tubulins, Tuba1a and Tubb4b, histone H2A, and filensin (Bfsp1), ranked #12, 16, 14, and 17, respectively. In contrast, crystallin proteins only made up 24% of the epithelium proteome, and several non-crystallin proteins, including histones H2A and H4, cytoplasmic actin, ubiquitin, and Aldh1a1, were within the top 10 protein ranks (#2, 7, 4, 8, and 9), respectively. Consistent with the finding that the mRNA levels of crystallin genes were lower in lens epithelium compared to fibers (see below), the most abundant crystallins in epithelium proteome, i.e. αA-, βB3-, βB1-, αB-, and βA1-crystallins were between 7.5% and 1.5% levels.
Given the key roles of crystallin proteins for lens transparency and their multifunctionality (Cvekl et al., 2015; Haslbeck and Vierling, 2015; Shiels and Hejtmancik, 2017; Slingsby et al., 2013; Thanos et al., 2014) and common transcriptional regulatory mechanisms governing their expression (Cvekl et al., 2017), the individual profiles of all crystallin mRNAs determined by RNA-Seq (Zhao et al., 2018) are shown in Fig. 4. These data show for the first time that expressions of the majority of crystallin mRNAs were significantly increasing from E14.5 to P0.5 in fiber (by adjusted p-value <0.05, in E16.5 VS E14.5, E18.5 VS E16.5 and P0.5 VS E18.5 comparisons), except for Cryab, Cryba1, Crybb2, Crygn, Crygs which significantly increased from E14.5 to E18.5 (Fig. 4). Our earlier studies of nascent mRNA transcription of Cryaa and Cryba1 in individual lens fiber cell nuclei showed that expression of these genes peaked at E16.5, while expression of Cryga reached maximal levels in central lens fiber cell nuclei approaching their destruction to form the organelle-free zone (Limi et al., 2018). These data quantitatively confirmed the well-established temporal pattern of crystallin gene expression in the lens fiber cell compartment, with αA- and αB- crystallins expressed at higher quantities than the other β-/γ-crystallins as their chaperone-like functions may be needed for proper folding and assembly of “late” β-/γ-crystallins in the lens fiber cell cytoplasm as well as for the formation of lens fiber cell cytoskeleton (Cheng et al., 2017; Song et al., 2009).
Fig. 4. Expression profiles of 15 individual crystallin genes by RNA-Seq.
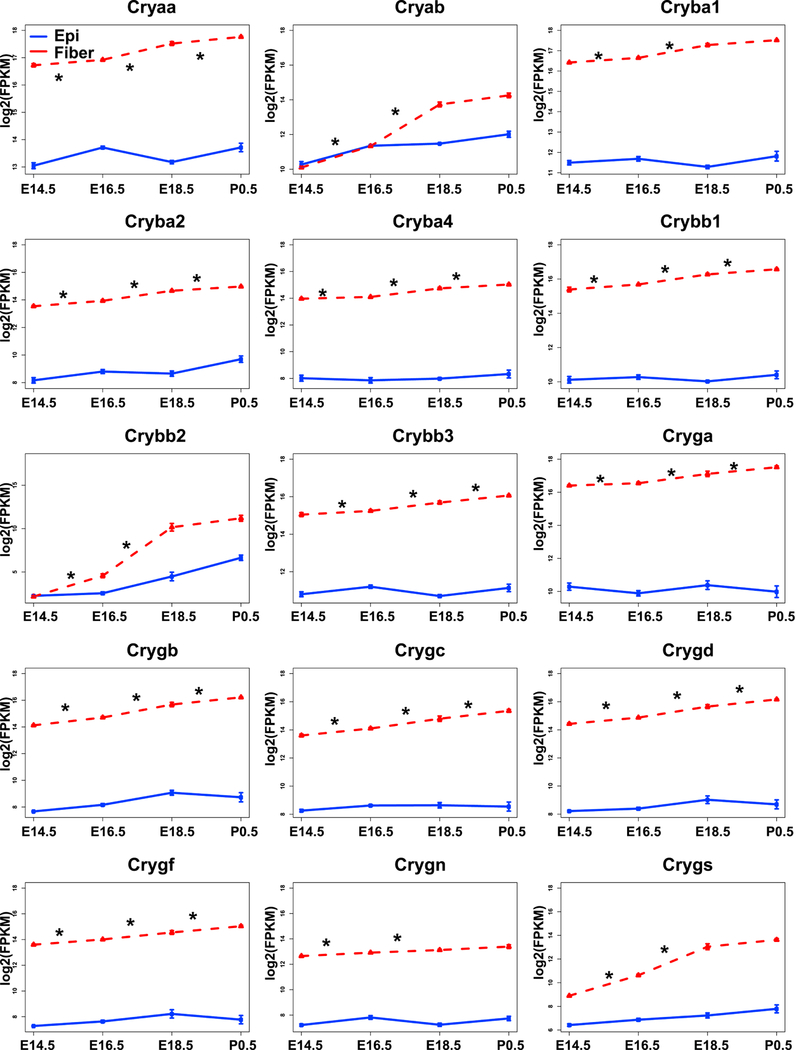
The graphs illustrate that expressions of most crystallins in lens fibers (red) increased from E14.5 to P0.5. The expression in epithelium (blue) is also shown. The “*” indicates adjusted p value (by Benjamini-Hochberg method) <0.05.
Expression of multiple crystallins was detected in laser microdissected mouse lens placodes (stages E9.5 and E10.5) by transcriptome profiling (Huang et al., 2011; Wolf et al., 2013). The quantitative order of individual transcripts is Cryaa > Crybb3 > Cryab > Cryge > Crygd > Crygb > Crygc > Cryba2 > Crygn in the invaginating lens placode. Previous studies established sequential appearance of Cryab and Cryaa mRNAs in the lens placode and lens pit, respectively (Robinson and Overbeek, 1996). Thus, low expression levels of β-/γ-crystallins in newborn lens epithelium found here were consistent with findings of multiple corresponding mRNAs in the invaginating lens placode (Huang et al., 2011; Wolf et al., 2013). Interestingly, proteomic analysis of the lens epithelium ranked all 15 major crystallins within range 1 to 449. Nevertheless, notable differences in their ranks between lens epithelium and lens fibers supported the high quality of our epithelial preparation.
It was well established that several proteins function as taxon-specific enzyme crystallins, including α-enolase (Eno1, Ƭ-crystallin, (Wistow et al., 1988)), aldehyde dehydrogenase family 1 subfamily 7A Aldh1a7 (η-crystallin, (Graham et al., 1996)), and lactate dehydrogenase A Ldha (ν-crystallin, (van Rheede et al., 2003)), in lamprey and turtle, elephant shrew, and platypus, respectively. Here we found that Ƭ-, η- and ν-crystallins were ranked #13, #145, #72 and #33, #385 and #238 in lens epithelium and fibers, respectively. Thus, we propose that expression dynamics of these genes will be useful markers for early developmental studies of the cells forming the lens pit and/or placode.
3.3. Analysis of DNA-binding transcription factors
The mRNAs encoding DNA-binding transcription factors were analyzed more thoroughly in the accompanying paper (Zhao et al., 2018). Here, we extended this analysis for the proteome (Fig. 5). Thirty-six transcription factors (21 have significantly higher mRNA levels in P0.5 lens epithelium), including FoxE3, Sp3, Onecut1, Creb1 and Tfcp2, were only found in the lens epithelium. In contrast, 11 transcription factors (5 have significantly higher mRNA levels in P0.5 lens fibers) were found only in lens fibers, including Pitx2, HoxB7, Hsf4, Mlxip and Nfx1. Finally, Ybx1, Tsc22d1, Ybx3, Gtf2i, Pax6 and Ubtf were found in both lens compartments. Lens defects have been previously reported in FoxE3, Pitx2, Hsf4, and Pax6 knockout mouse models (Collinson et al., 2001; Fujimoto et al., 2004; Kelberman et al., 2011; Medina-Martinez et al., 2005; van Raamsdonk and Tilghman, 2000). Pax6 mouse heterozygous lenses show completely disrupted crystallin gene expression profile (Xie and Cvekl, 2011) while Hsf4 null lenses mostly disrupt expression of γ-crystallins (Fujimoto et al., 2004) and HSF4 cataract causing missense mutation reduces expression of αB-crystallin in an animal model (Jing et al., 2014). Amongst the signal-regulated transcription factors, the present proteomic analysis identified three, including Creb1 (Cvekl et al., 1995; Yang et al., 2006), Smad4 (Liu et al., 2010), and Stat3 (Potts et al., 1998) that mediate canonical cAMP, BMP/TGF-β, and JAK/STAT signaling in the vertebrate lens, respectively. Note that Ybx1 (Shurtleff et al., 2016; van Zalen et al., 2015) and Ybx3 (Snyder et al., 2015) also function as RNA-binding proteins and regulators of translation and as sorters of specific microRNAs.
Fig. 5. Proteomic analysis of DNA-binding transcription factors.
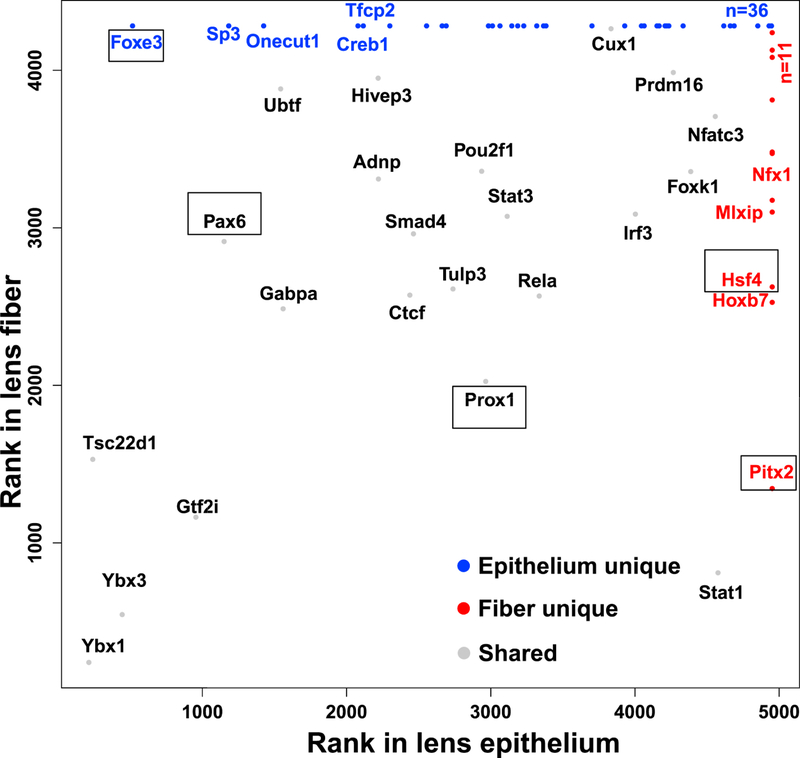
The graph shows three groups of transcription factors: found in both lens compartments (grey), only in lens epithelium (blue), and only in lens fibers (red). Transcription factors studied by loss-of-function studies and regulating lens formation are boxed.
3.4. Analysis of RNA-binding proteins
Analysis of RNA-binding proteins within the epithelium vs. fiber-enriched groups revealed major differences between proteins found in the lens epithelium (n=49) and those in lens fibers (n=27) (Fig. 2; Table S1). These RNA-binding proteins are involved in RNA splicing, regulation of mRNA translation and stability, and other cellular functions. Consistent with previous studies showing that fiber cell abundant proteins Caprin2 (Enrichment ratio=18.2) (Dash et al., 2015) and Tdrd7 (Enrichment ratio=11.3) (Lachke et al., 2011) played major roles in lens morphogenesis, both Caprin2 (#156) and Tdrd7 (#242) belonged to the most highly abundant fiber cell proteins and were enriched in fiber together with polyadenylate-binding proteins 1 and 2 (Pabpc1/2) (#251, Enrichment ratio=2.3), and Caprin1 (#257, Enrichment ratio=2.2) (Fig. 7B). Increased abundance of RNA-binding Motif protein 38 (Rbm38, #341, Enrichment ratio=14.5) and Purkinje cell protein 4 (Pcp4, #357, Enrichment ratio=3.1) in lens fibers mimicked trends found with Caprin2 and Tdrd7, making these proteins excellent candidates for future functional studies.
Fiber-cell-abundant protein Rbm38 regulates splicing during late erythroid differentiation (Heinicke et al., 2013) and the lens might use similar mechanisms. In contrast, CUGBP Elav-like family member 1 (Celf1) protein only ranked #1746 in lens fibers yet a recent study shows its critical role in posttranscriptional regulation of Cdkn1b/p27 and Dnase2b mRNAs (Siddam et al., 2018). Rbm3 (Enrichment ratio=4.3) is abundant both in lens epithelium (#91) and lens fibers (#390) (Table S1). Mutations in human RBM3 are associated with high myopia and retinal dystrophy (Arno et al., 2015). Pabpc1/2 proteins bind to poly(A) to facilitate ribosome recruitment, translational initiation, and are also required for initial steps of mRNA decay (Stupfler et al., 2016). Both Hnrnpk (Enrichment ratio=2.2) and Hnrnph1 (Enrichment ratio=1.8) were co-immunoprecipitated with Pax6 in lens nuclear extracts (Sun et al., 2016) and influence pre-mRNA processing expression of heat shock proteins Hsp27 and Hsp70 (Kim et al., 2017). Another trend was epithelial-preferred expression of several serine/arginine rich splicing factors, including Srsf1 (Enrichment ratio=3.9), Srsf7 (Enrichment ratio=2.6), Srsf5 (Enrichment ratio=2.9), and Srsf10 (Enrichment ratio=2.8), that might be selectively employed to control alternate splicing between lens epithelial and fiber cells (Srivastava et al., 2017). A cold-shock domain-containing calcium regulated heat stable protein 1 (Carhsp1, also called Crhsp24, Enrichment ratio=3.4) is a cytoplasmic protein used to stabilize specific mRNAs (Hou et al., 2011). It is localized to processing bodies (P-bodies) and cytoplasmic exosomes but not on translating mRNAs (Pfeiffer et al., 2011) in mouse leukemic monocyte-macrophage cell line. Carhsp1 is one of the mostly highly abundant lens RNA-binding proteins (epithelium ranking #244, fiber ranking #72). Our localization studies identified this protein in the cytoplasm at the apical edges of the lens fibers as well as in lens epithelium. Carhsp1 proteins were also found in the cell nuclei in lens epithelium and peripheral fibers as well as in the anterior regions of lens fiber but mostly in the cytoplasm (Fig. 6). These staining patterns support the idea that Carhsp1 regulates multiple stages of the mRNA cycle of abundant lens transcripts. Ongoing experiments are aimed to identify mRNAs bound to Carhsp1.
Fig. 6. Immunofluorescence analysis of expression of Carhsp1 in mouse lens.
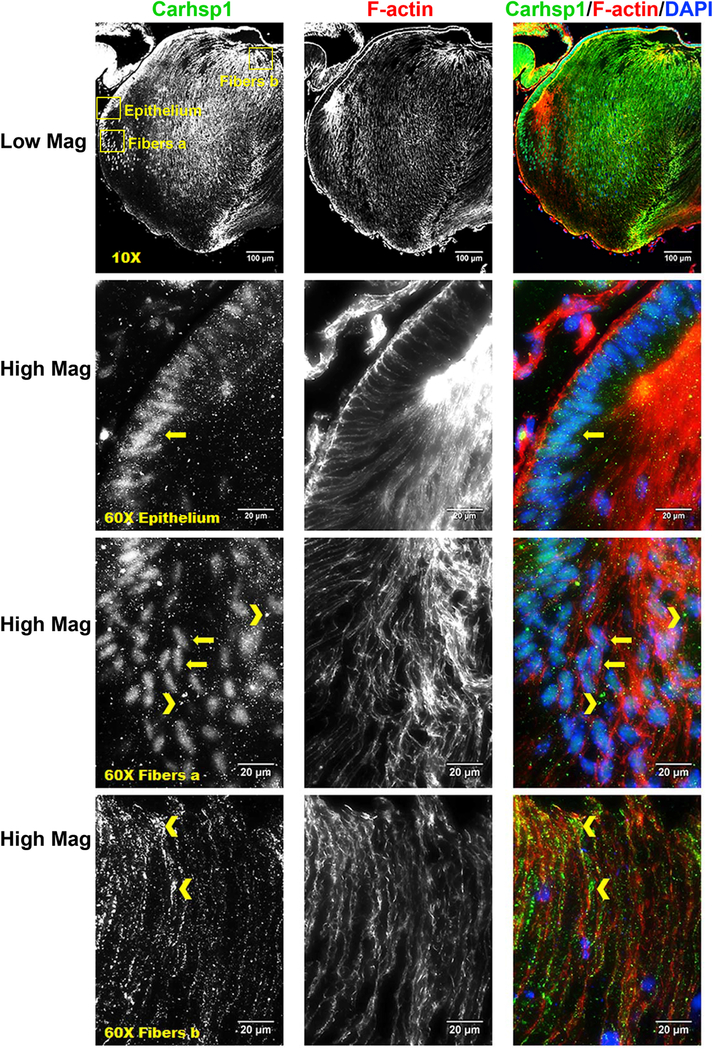
Immunostaining for Carhsp1 (green), F-actin (red) and DAPI (nuclei, blue) in P0.5 CD1 mouse lens sections. Low magnification images show Carhsp1 is expressed in both lens epithelium and fiber. High magnification images demonstrate Carhsp1 is mostly found in the nuclei at lens epithelial as well as periphery fiber cells (see yellow arrows) while in the cytoplasm (see yellow chevrons) at anterior (apical) portions of fiber cells. Yellow boxes were used to indicate the positions of high magnification images, including epithelium, fibers position a (periphery fiber cells), and fibers position b (anterior/apical fiber cells). Scale bars: 100 µm (low magnification, top panels) and 20 µm (high mag, lower panels).
3.5. Formation of lens fiber cell cytoskeleton and microtubules
Lens fiber cell cytoskeleton is comprised of universal and lens-specific protein components (Cheng et al., 2017; Rao and Maddala, 2006; Song et al., 2009). A large number of the cytoskeletal proteins identified by our screen have known localization and functions in the lens. Actin-associated periaxin (Prx, Enrichment ratio=10.4) is highly enriched in lens fiber cells of the developing lens, and loss of Prx leads to a mispacking of hexagonal lens fiber cells (Maddala et al., 2011b). Both non-muscle myosin IIA (Myh9, rank#122, Enrichment ratio=4.4) and IIB (Myh10, rank #273, Enrichment ratio=3.2) were found abundant in the lens epithelium while related myosin IIC (Myh14) was significantly less expressed (rank #3340). Expression of both these myosin proteins in the epithelium suggests that the contractile actomyosin network may be important for lens epithelium function and integrity. Immunostaining revealed that myosin IIA was found mostly in the epithelial cells and is highly concentrated along the apical-apical interface between epithelial and fiber cells (Fig. 8A). Mutations in MYH9 cause human cataracts and other diseases, such as macrothrombocyopenia, nephropathy and sensorineural hearing loss (Aoki et al., 2018; De Rocco et al., 2013). Myosin IIB (Myh10, Enrichment ratio=3.2) is important for active contraction of F-actin networks. Myosin IIB is highly expressed in lens placode epithelial cells and is needed for normal placode invagination (Chauhan et al., 2009; Plageman et al., 2010). This is consistent with the epithelial-cell-enrichment we observed for Myh10 in the proteome.
Fig. 8. Immunofluorescence analysis of expression of Myh9 and Sptb in mouse lens sections.
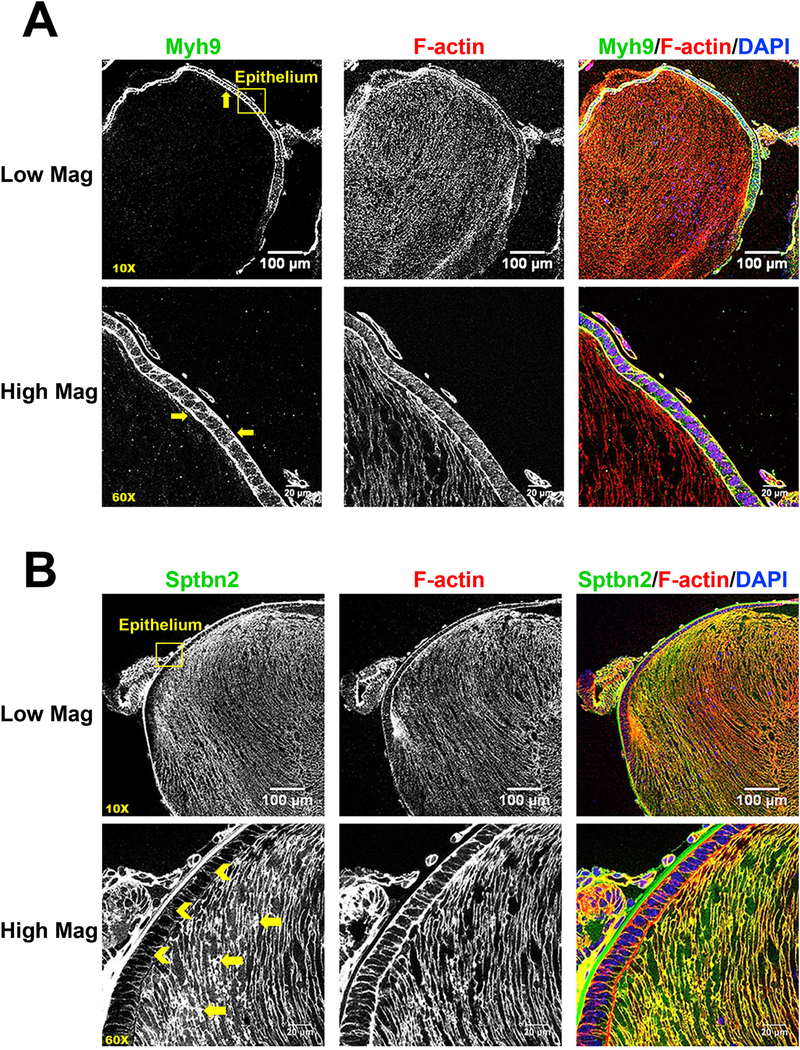
A) Immunostaining for Myh9 (green), F-actin (red) and DAPI (nuclei, blue) in P0.5 CD1 mouse lens sections. Low magnification (10X) images show that Myh9 is expressed mostly in lens epithelial cells (see yellow arrows). High magnification (60X) images demonstrate the Myh9 is highly concentrated at the apical-apical interface between lens epithelial and fiber cells (see yellow arrows). B) Immunostaining for β2-spectrin (green), F-actin (red) and DAPI (blue) in mouse lens sections reveals that β2-spectrin is highly expressed in lens fiber cells (see yellow arrows) with low signal in lens epithelial cells (see yellow chevrons). Yellow boxes were used to indicate the positions of high magnification images. Scale bars: 100 µm (low magnification, top panels) and 20 µm (high mag, lower panels).
Three spectrin proteins, Sptb, Sptbn1 and Sptbn2, were detected predominantly in the lens fiber proteome (Sptbn1 > Sptbn2 >> Sptb). Non-erythrocyte β2-spectrin (Sptbn2, Enrichment ratio=4.4) is important for the formation of the membrane skeleton with tropomodulin 1 (Tmod1)-capped F-actin in the lens (Cheng et al., 2016; Gokhin et al., 2012; Nowak et al., 2009; Woo et al., 2000) (Fig. 8B). Consistent with this proteomics data, Tmod1 (Enrichment ratio=11.0) is highly expressed and localized to fiber cell membranes, but have low expression in epithelial cells (Nowak et al., 2009). In agreement with the proteomics results, our immunostaining data reveals that β2-spectrin (Enrichment ratio=4.3) is highly expressed in lens fiber cells, with lower staining signal in epithelial cells (Fig. 8B).
The actin cytoskeleton is likely very dynamic in both epithelium and fiber due to the high abundance of cofilin-1 (Cfl1, ranked #62 in epithelial cells and #116 in fibers, Enrichment ratio=1.9), profilin-1 (Pfn1, ranked #60 in epithelial cells and #64 in fibers, Enrichment ratio=1.1) and gelsolin (Gsn, ranked #247 in epithelial cells and ranked # 78 in fibers, Enrichment ratio=3.2). These three proteins are important for assembly and dynamic remodeling of F-actin networks (Skruber et al., 2018). Cfl1 is an F-actin severing protein (Ostrowska and Moraczewska, 2017) while Pfn1 binds and sequesters G-actin monomers and catalyzes F-actin assembly in a concentration dependent manner (Alkam et al., 2017). Gsn is able to cap and sever F-actin and can also sequester G-actin (Nag et al., 2013).The balance of these actin sequestering, severing and polymerization factors are likely needed to remodel the F-actin network as epithelial cells divide, differentiate and elongate into fiber cells and as fiber cell membrane morphology increases in complexity during cell maturation (Cheng et al., 2016; Dickson and Crock, 1972; Kistler et al., 1986; Kuszak et al., 1980; Kuwabara, 1975; Lo and Harding, 1984; Willekens and Vrensen, 1981, 1985). Rac proteins (Enrichment ratio=2.0) were present in both epithelium and fiber compartments, with no enrichment in either. Regulation of F-actin by Rac family small GTPase 1 (Rac1) is required for normal lens shape, maintenance of lens epithelial cells and fiber cell orientation (Maddala et al., 2011a).
Lens fiber cells also form a specialized beaded intermediate filament network comprised of lens-specific beaded filament structural proteins 1 and 2, Bfsp1 (filensin, Enrichment ratio=21.5) and Bfsp2 (phakinin or CP49, Enrichment ratio=21.3) (Alizadeh et al., 2003; Sandilands et al., 1995b). Previous studies showed that CP49 and filensin had enhanced immunostaining signal in lens fiber cells (Ireland et al., 2000; Simirskii et al., 2006; Sun et al., 2015a), consistent with fiber-cell-enrichment in our dataset. Beaded intermediate filaments are important for maintaining the integrity of the innermost mature fiber cells (Yoon et al., 2008) and for lens biomechanical properties (Fudge et al., 2011; Gokhin et al., 2012). In contrast, lens epithelial cells have a rich vimentin network (Song et al., 2009; Sun et al., 2015a), consistent with our data showing high expression of vimentin (rank #24, Enrichment ratio=1.1). Overexpression of vimentin in the lens leads to abnormal vacuoles in newly differentiating fiber cells and inner fiber cells degeneration (Capetanaki et al., 1989), suggesting that too much vimentin inhibits the normal differentiation programming from epithelial cells to fiber cells. These ranking differences implicated different needs of lens fiber cells during their morphogenesis that was achieved by proteome remodeling, including the formation of intermediate filaments from lens-specific proteins Bfsp1 and Bfsp2, and vimentin (Perng et al., 2007; Sandilands et al., 1995a; Song et al., 2009).
Lens epithelial and fiber cells contained abundant tubulin isoforms forming microtubules, including α1-, α3-, β4b-, β5-, β1b-, and β2a-tubulins (Enrichment ratios range from 1.0–1.9) (Table S1). There was no obvious enrichment of tubulin subunits in either cell compartment, other than β6-tubulin enriched in fiber. The α-tubulin data agreed with a recent work showing ubiquitous α-tubulin staining in embryonic mouse lens sections (Logan et al., 2018).
3.6. Analysis of membrane proteins and its limitation
Despite the expected negative bias towards detection of membrane proteins in typical shotgun proteomics protocols, our data revealed 29 lens integral membrane and membrane-associated proteins and 12 cell adhesion proteins (Table S1). The top protein components of lens fiber cell membranes were enriched for Mip (aquaporin 0, Enrichment score=36.3) and Lim2 (MP20, Enrichment score=10.0). Earlier studies linked these proteins and their encoded genes with lens membrane structure (Gorin et al., 1984; Maher et al., 2012; Mulders et al., 1995) and identified cataract-causing mutations (Irum et al., 2016; Shi et al., 2011; Shiels et al., 2007). Recent studies have shown that Mip was important for fiber cell integrity (Bennett et al., 2016), lens biomechanical properties (Sindhu Kumari et al., 2015) and ion and fluid homeostasis (Kumari et al., 2017), while Lim2 was required for fiber cell adhesion (Shi et al., 2011). Mip and Lim2 staining patterns were similarly increased in lens fiber cells with membrane insertion in inner mature fiber cells (Bassnett et al., 2011; Schey et al., 2017) consistent with our enrichment analysis. Interestingly, aquaporin 1 (Aqp1, Enrichment ratio=9.2) was enriched in the epithelium, consistent with its known staining pattern in the lens and its function in regulating water permeability in the lens (Schey et al., 2017). In contrast to earlier studies of lens membrane proteins (Bassnett et al., 2009; Grey et al., 2013; Petrova et al., 2015), aquaporin 5 (Aqp5) was not detected in current samples, which was consistent with its low expression levels in our RNA-seq data (Zhao et al., 2018) using CD1 mice as well as previously published RNA-seq data using FVB mice (Hoang et al., 2014).
Consistent with previous work, other top fiber-cell-enriched proteins included gap junction protein 3 (Gja3, connexin 46, Enrichment score=6.1) (Gong et al., 1997), gap junction protein 8 (Gja8, connexin 50, Enrichment score=3.2) (Graw, 2009; Rong et al., 2002; Wang et al., 2016), griffin (Enrichment score=4.0) (Ogden et al., 1998), brain acid soluble protein 1 (Basp1, Enrichment score=2.6) (Bagchi et al., 2008; Wenke et al., 2016), calcium regulator Slc3a2 (Enrichment score=1.5) (Bassnett et al., 2009), and glucose transporter Slc2a1 (Glut1, Enrichment score=3.0) (Swarup et al., 2018). Connexins 46 and 50 were highly expressed in lens fiber cells and were required for lens transparency and growth (Gong et al., 2007). These proteins formed important large gap junction plaques between lens fiber cells to facilitate the outflow pathway of the lens microcirculatory pathway (Cheng et al., 2015; Mathias et al., 2010).
The most notable differences in cell adhesion molecules were epithelium-preferred fibronectin 1 (Fn1, Enrichment score=9.1) (Hayes et al., 2012) and neural cell adhesion 1 (Ncam1, Enrichment score=2.8) (Bassnett et al., 2009). In contrast, armadillo repeat gene deleted in velocardifacial syndrome (Arvcf, Enrichment score=2.5 (Bassnett et al., 2009; Wistow et al., 2002), α2-catenin (Ctnna2, Enrichment score=5.0 (Bassnett et al., 2009), and neuronal cell adhesion molecule (Nrcam, Enrichment score=6.7 (More et al., 2001), were more prominent in the fiber cell proteome (Table S1). In terms of membrane remodeling, our data pointed to a much lower ranking of basigin (Bsg, Enrichment score=4.9), vesicle-associated membrane protein 2 (Vamp2, Enrichment score=1.8), aquaporin-1 (Aqp1, Enrichment score=9.2), and carbonic anhydrase 14 (Ca14, Enrichment score=4.1) in fiber cell vs epithelial cell proteome pointing to their important functions in the lens epithelial cells. Recent studies outside of lens showed that basigin is an integral partner of plasma membrane Ca2+-ATPases, key regulators of Ca2+ signaling (Schmidt et al., 2017), and Vamp2 is a component of synaptic vesicles (Lou et al., 2017). While no specific role of Ca14 is known in the lens, fluid transport is critical for maintenance of lens homeostasis (Mathias et al., 2010; Paterson and Delamere, 2004).
3.7. Analysis of ribosomal subunits and translational system
Lens fiber cell differentiation requires robust production of high-quality proteins that are essential for refractive index and transparency. Since degradation of fiber cell nuclei and endoplasmic reticulum during cell maturation imposed additional restrictions on the translational system, we examined the distribution of individual ribosomal proteins (Yoshihama et al., 2002) amongst the top 525 proteins between lens epithelium and lens fibers (Fig. 9). We found that several ribosomal subunits, including Rps10, Rpl23a, Rps3a, Rps16, Rpl13, Rpl7a, Rps25, Rpl18a, Rpl36, Rpl10, Rpl10l, and Rps12 (Enrichment scores range from 2.0 to 11.8), were enriched in the fiber cell proteome. In contrast, the lens epithelium showed more abundant Rps27a and Rps27l (Enrichment score=3.1 and 3.5, respectively). Rps27a is produced as an N-terminal ubiquitin and C-terminal small ribosomal protein fusion (Redman and Rechsteiner, 1989), and Rps27l is 96% identical to Rps27 and also plays a regulatory role in apoptosis (He and Sun, 2007). These findings raised the intriguing possibility that both lens compartments employed specialized ribosomes (Briggs and Dinman, 2017) that optimally produce crystallins and other abundant lens proteins. Importantly, reduced expression of RPL7A, RPL13A, RPL15, and RPL21 were found in human age-related cataract samples (Zhang et al., 2002) supporting the idea that specialized ribosomes indeed function in the lens.
Fig. 9. Proteomic analysis of small and large ribosomal subunits.
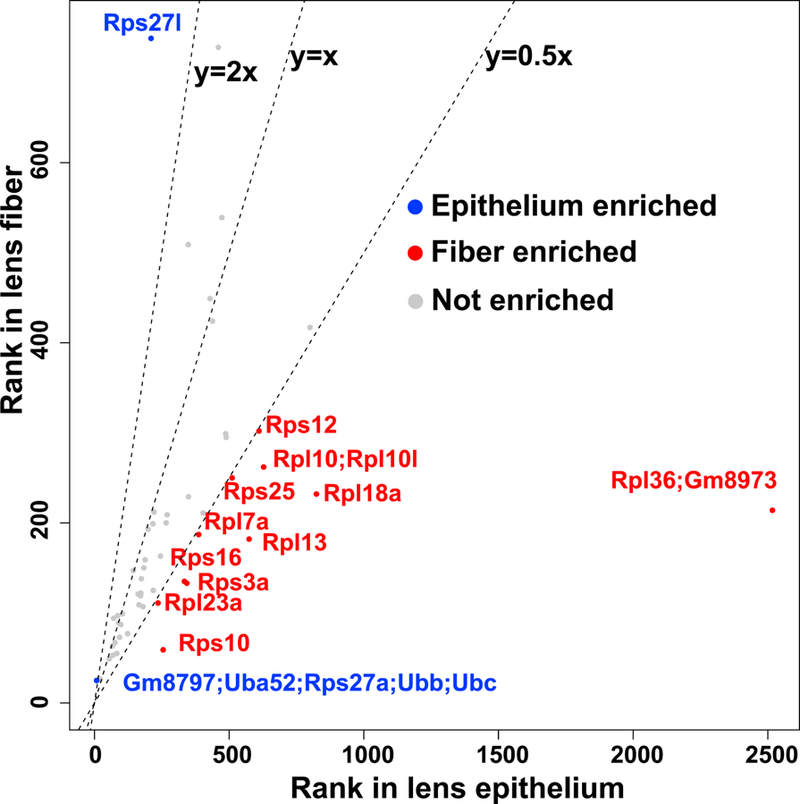
Ribosomal protein ranks in lens epithelium and lens fiber are shown. A small ribosome 40S subunit, Rps; a large ribosome 60S subunit, Rpl.
Twenty initiation factors of translation, subunits of the eIF1-eIF6 complexes (Enrichment ratios range from 1.2 to 3.4), were ranked within the top 525 proteins and also showed notable differences between lens epithelium and fibers (Fig. 7, Table S1). The common trend was an increased abundance of 10 of these subunits in the fibers to support the translational machinery (Fig. 7). Interestingly, the 3’-UTRs of multiple crystallin-mRNAs in zebrafish were recognized by a subunit eIF3h (Choudhuri et al., 2013) which was found outside of the top 525 group (rank #1,660 in epithelium and #1,188 in fiber proteomes, respectively). On the other hand, there was a preference to express the “non-core” subunits (Hinnebusch, 2006; Siridechadilok et al., 2005), including eIF3k, eIF3f, eIF3l, eIF3m, and eIF3d (Enrichment ratios range from 1.4 to 3.4), in lens fibers. Both translation and transcript stability are regulated by fiber cell-enriched eIF4e (Enrichment ratio=2.1), Pabpc1 and Pabpc6 (Enrichment ratio=2.4), and Eif3g (Enrichment ratio=2.0). These proteins form the “mRNA closed loop” structure together that regulates both translation and mRNA stability (Jackson et al., 2010). It is thus possible that these fiber-enriched subunits control translation of crystallins and other abundant mRNAs in lens fibers and that many proteins discussed earlier in section 3.4 are also involved in mRNA translation and stability.
Fig. 7. Proteomic analysis of RNA-binding, microtubule, cytoskeletal, membrane and initiation factor proteins.
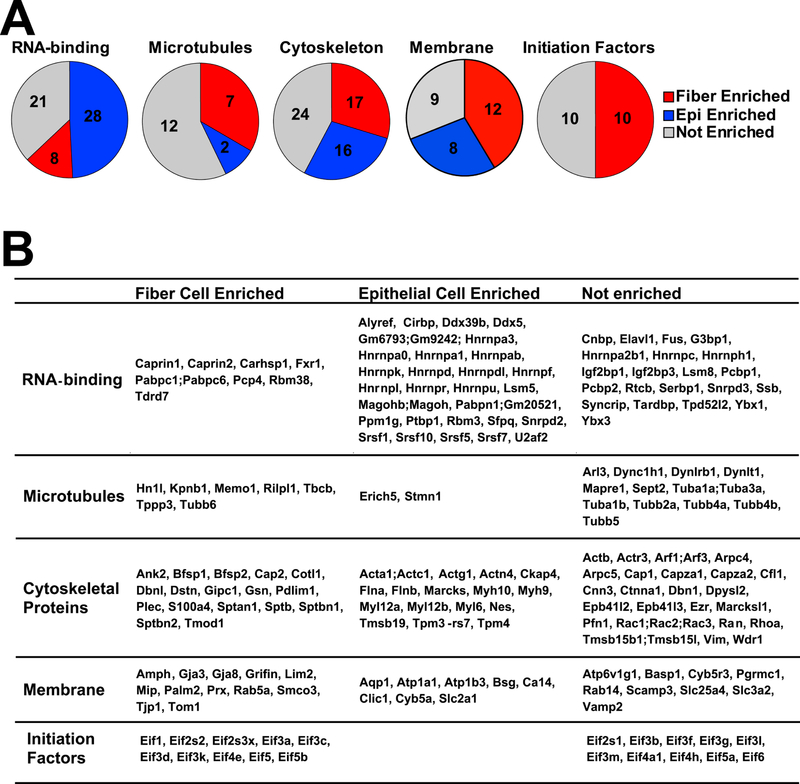
(A). Pie charts show numbers of epithelium-, fiber- and not enriched proteins inside each group. (B). The table shows the enriched and not enriched proteins alphabetically listed inside each group.
3.8. Lens epithelial and fiber cell phenotypes
Systematic analysis of all proteins, RNAs, and other molecules that collectively contribute to specific cell types is challenging due to natural complexity of individual tissues; nevertheless, it is now becoming feasible due to advances in methods applicable at single cell levels (Buenrostro et al., 2015; Macosko et al., 2015). Vectors of cellular identity as well underlying organization of the chromatin and nuclei have to be identified (Wagner et al., 2016). While we have excellent lens-specific markers, including crystallins, filensin, Lim2, Mip, and phakinin, additional proteins need to be identified for specific stages and subcellular compartments of the developing and mature lens.
Lens formation can be viewed as a series of cell fate decisions including the formation of lens progenitor cells, precursor cells, nascent epithelium and fibers, and, finally, mature epithelium and fibers. Nevertheless, different populations of cells exist within these compartments, such as quiescent anterior vs. proliferative equatorial epithelial cells and fibers at different stages of differentiation and maturation before and after organelle degradation (Bassnett and Sikic, 2017; Cvekl and Ashery-Padan, 2014). Unfortunately, with micro-dissection method, we could not resolve the proteome changes during epithelial cell division and fiber cell organelle degradation. Additionally, due to the sonication and tissue grinding, some membrane proteins might not be detected as described above. However, our data provided novel insights into the abundance of lens structural proteins between lens epithelium and fibers, and expression of these genes can be traced back during the formation of lens progenitor cells to understand their presumptive heterogeneity and plasticity. For example, two enzymes, Aldh1a1 and Eno1, were amongst the top 10 most highly expressed proteins in the lens epithelium and it will be interesting to track their expression during the earliest stages of lens cell formation. High expression of αA-, βB3-, and αB-crystallins in newborn lens epithelium correlated with their early expression in the lens placodes (Huang et al., 2011; Wolf et al., 2013). Comparative analysis of their mRNAs together with DNA-binding transcription factors at single cell level will generate new testable hypotheses to reconstruct gene regulatory networks underlying early stages of lens induction. Likewise, transcriptional regulation of lens fiber-enriched non-crystallin encoding genes remains in its infancy, and current data will assist in prioritization of these studies. Finally, functions of numerous abundant proteins in lens fibers, including α4-actinin (Actn4), coactosin-like protein (Cotl1), α-and β-spectrins, protein S100A4, plectin (Plec), Pdlim1, and others, remain unknown or poorly understood. The enrichment analysis also revealed new markers for epithelial vs. fiber cells that will be important benchmarks for in vitro lens-like organoid cultures to verify segregated cell populations that mimic lens development.
4. Conclusion
The present study is the first combined analysis of unique lens epithelial and lens fibers proteomes and transcriptomes. We show that analysis of as few as 525 most abundant proteins within each lens compartment generated novel information regarding critical processes involved in lens formation and was linked to distinct cellular and molecular organization of lens epithelial and lens fiber cells. These lens proteins can be grouped into eight groups, including nucleus and DNA, protein synthesis, membrane, cytoskeleton, signaling, mitochondria, degradation and recycling, and metabolism. Although the functions of a number of these proteins are already known in the lens, functions of the majority of these proteins in the lens remain unknown. Quantitative analysis of individual proteins with unknown roles in the lens together with their roles outside of the lens provides a clear rationale to prioritize the follow-up studies including analysis of subcellular localization, interactions between proteins and DNA/RNA polymers, and mapping of protein-protein interactions.
Although the proteomics data are from three large-scale analyses of pooled lens epithelium, lens fibers, and whole lens performed without replication, the reproducibility of these experiments are supported by the parallel analyses of transcriptomes by RNA-seq conducted in triplicate, by comparison of the whole lens proteome to each region of the lens, and by analyses using the top abundance ranked proteins. Future studies are planned to directly address reproducibility, including duplicate analysis of lenses from C57Bl6 and FVB strains with stable isotope labeling (tandem mass tagging) to determine biological and technical variation. The full proteomics results and datasets produced in this study provide additional opportunities to explore the 4,000 lens proteins that were not discussed in detail here. It is important to point out that several well-known lens membrane proteins such as E-cadherin (Cdh1), Ephrin-B2 (Efnb2), Ephrin-A5 (Epha5), Ephrin type-B receptor 2 (Ephb2), and gap junction α−1 protein/connexin 43 (Gja1), were likely found outside of the top 525 most abundant proteins due to the experimental limitations.
Our study took a conservative approach to explore relationships between mRNA expression and protein expression levels; nevertheless, lens crystallins showed a high correlation between their mRNA and protein levels. Changes in protein ranking between lens epithelium and lens fibers supported a model of proteome remodeling that drives lens fiber cell morphogenesis and resulting intricate microarchitecture of lens fiber cells. Taken together, this work identified and prioritized proteins for future lens localization, protein-protein interactions, and functional studies to understand their roles in lens morphogenesis, protein synthesis quality control, and transparency.
Supplementary Material
Highlights:
First comparative analysis of mouse lens proteomes and transcriptomes
Reveals quantitative differences in proteins found in lens epithelium and lens fibers
Identifies fiber cell enriched protein components of the translational system
Evidence for proteome remodeling in differentiating lens fiber cells
Data source for prioritizing protein candidates for functional studies
Acknowledgement
Funding: NIH R01 EY012200 (AC), EY014237 (AC), P30 EY010572 (LLD), R01 EY027768 (LLD), R01 EY017724 (VMF), and R21 EY027389 (CC). We would like to thank Hillary Guzik, MSc for expert advice/assistance in microscopy. The imaging was conducted in the Analytical Imaging Facility partially funded by the NCI Cancer Grant P30CA013330. We also thank Dr. Salil Lachke and Dr. Carolina Eliscovich for their critical comments on analysis of RNA-binding proteins.
Abbreviations:
- eIFs
elongation initiation factors
- ECM
extracellular matrix proteins
- GO
Gene Ontology
- 2D-LC/MS
tandem mass spectrometry
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alizadeh A, Clark J, Seeberger T, Hess J, Blankenship T, FitzGerald PG, 2003. Targeted deletion of the lens fiber cell-specific intermediate filament protein filensin. Invest Ophthalmol Vis Sci 44, 5252–5258. [DOI] [PubMed] [Google Scholar]
- Alkam D, Feldman EZ, Singh A, Kiaei M, 2017. Profilin1 biology and its mutation, actin(g) in disease. Cellular and molecular life sciences : CMLS 74, 967–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki T, Kunishima S, Yamashita Y, Minamitani K, Ota S, 2018. Macrothrombocytopenia With Congenital Bilateral Cataracts: A Phenotype of MYH9 Disorder With Exon 24 Indel Mutations. Journal of pediatric hematology/oncology 40, 76–78. [DOI] [PubMed] [Google Scholar]
- Arno G, Hull S, Robson AG, Holder GE, Cheetham ME, Webster AR, Plagnol V, Moore AT, 2015. Lack of Interphotoreceptor Retinoid Binding Protein Caused by Homozygous Mutation of RBP3 Is Associated With High Myopia and Retinal Dystrophy. Invest Ophthalmol Vis Sci 56, 2358–2365. [DOI] [PubMed] [Google Scholar]
- Audette DS, Anand D, So T, Rubenstein TB, Lachke SA, Lovicu FJ, Duncan MK, 2016. Prox1 and fibroblast growth factor receptors form a novel regulatory loop controlling lens fiber differentiation and gene expression. Development 143, 318–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagchi M, Kousis S, Maisel H, 2008. BASP1 in the lens. Journal of cellular biochemistry 105, 699–702. [DOI] [PubMed] [Google Scholar]
- Barnes S, Quinlan RA, 2017. Small molecules, both dietary and endogenous, influence the onset of lens cataracts. Exp Eye Res 156, 87–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassnett S, Shi Y, Vrensen GF, 2011. Biological glass: structural determinants of eye lens transparency. Philos Trans R Soc Lond B Biol Sci 366, 1250–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassnett S, Sikic H, 2017. The lens growth process. Prog Retin Eye Res 60, 181–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassnett S, Wilmarth PA, David LL, 2009. The membrane proteome of the mouse lens fiber cell. Mol Vis 15, 2448–2463. [PMC free article] [PubMed] [Google Scholar]
- Bennett TM, Zhou Y, Shiels A, 2016. Lens transcriptome profile during cataract development in Mip-null mice. Biochemical and biophysical research communications 478, 988–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs JW, Dinman JD, 2017. Subtractional Heterogeneity: A Crucial Step toward Defining Specialized Ribosomes. Mol Cell 67, 3–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, Chang HY, Greenleaf WJ, 2015. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capetanaki Y, Smith S, Heath JP, 1989. Overexpression of the vimentin gene in transgenic mice inhibits normal lens cell differentiation. J Cell Biol 109, 1653–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalheiro GR, Matos-Rodrigues GE, Zhao Y, Gomes AL, Anand D, Predes D, de Lima S, Abreu JG, Zheng D, Lachke SA, Cvekl A, Martins RAP, 2017. N-myc regulates growth and fiber cell differentiation in lens development. Dev Biol 429, 105–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan BK, Disanza A, Choi SY, Faber SC, Lou M, Beggs HE, Scita G, Zheng Y, Lang RA, 2009. Cdc42- and IRSp53-dependent contractile filopodia tether presumptive lens and retina to coordinate epithelial invagination. Development 136, 3657–3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Sagar V, Len HS, Peterson K, Fan J, Mishra S, McMurtry J, Wilmarth PA, David LL, Wistow G, 2016. gamma-Crystallins of the chicken lens: remnants of an ancient vertebrate gene family in birds. Febs j 283, 1516–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Nowak RB, Biswas SK, Lo WK, FitzGerald PG, Fowler VM, 2016. Tropomodulin 1 Regulation of Actin Is Required for the Formation of Large Paddle Protrusions Between Mature Lens Fiber Cells. Invest Ophthalmol Vis Sci 57, 4084–4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Nowak RB, Fowler VM, 2017. The lens actin filament cytoskeleton: Diverse structures for complex functions. Exp Eye Res 156, 58–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Nowak RB, Gao J, Sun X, Biswas SK, Lo WK, Mathias RT, Fowler VM, 2015. Lens ion homeostasis relies on the assembly and/or stability of large connexin 46 gap junction plaques on the broad sides of differentiating fiber cells. American journal of physiology. Cell physiology 308, C835–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhuri A, Maitra U, Evans T, 2013. Translation initiation factor eIF3h targets specific transcripts to polysomes during embryogenesis. Proceedings of the National Academy of Sciences of the United States of America 110, 9818–9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinson JM, Quinn JC, Buchanan MA, Kaufman MH, Wedden SE, West JD, Hill RE, 2001. Primary defects in the lens underlie complex anterior segment abnormalities of the Pax6 heterozygous eye. Proceedings of the National Academy of Sciences of the United States of America 98, 9688–9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M, 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature biotechnology 26, 1367–1372. [DOI] [PubMed] [Google Scholar]
- Cvekl A, Ashery-Padan R, 2014. The cellular and molecular mechanisms of vertebrate lens development. Development 141, 4432–4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvekl A, Kashanchi F, Sax CM, Brady JN, Piatigorsky J, 1995. Transcriptional regulation of the mouse alpha A-crystallin gene: activation dependent on a cyclic AMP-responsive element (DE1/CRE) and a Pax-6-binding site. Molecular and cellular biology 15, 653–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvekl A, McGreal R, Liu W, 2015. Lens Development and Crystallin Gene Expression. Prog Mol Biol Transl Sci 134, 129–167. [DOI] [PubMed] [Google Scholar]
- Cvekl A, Zhang X, 2017. Signaling and Gene Regulatory Networks in Mammalian Lens Development. Trends Genet 33, 677–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvekl A, Zhao Y, McGreal R, Xie Q, Gu X, Zheng D, 2017. Evolutionary Origins of Pax6 Control of Crystallin Genes. Genome biology and evolution 9, 2075–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash S, Dang CA, Beebe DC, Lachke SA, 2015. Deficiency of the RNA binding protein caprin2 causes lens defects and features of Peters anomaly. Developmental dynamics : an official publication of the American Association of Anatomists 244, 1313–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rocco D, Zieger B, Platokouki H, Heller PG, Pastore A, Bottega R, Noris P, Barozzi S, Glembotsky AC, Pergantou H, Balduini CL, Savoia A, Pecci A, 2013. MYH9-related disease: five novel mutations expanding the spectrum of causative mutations and confirming genotype/phenotype correlations. Eur J Med Genet 56, 7–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson DH, Crock GW, 1972. Interlocking patterns on primate lens fibers. Investigative ophthalmology 11, 809–815. [PubMed] [Google Scholar]
- Fu Q, Qin Z, Jin X, Zhang L, Chen Z, He J, Ji J, Yao K, 2017. Generation of Functional Lentoid Bodies From Human Induced Pluripotent Stem Cells Derived From Urinary Cells. Invest Ophthalmol Vis Sci 58, 517–527. [DOI] [PubMed] [Google Scholar]
- Fudge DS, McCuaig JV, Van Stralen S, Hess JF, Wang H, Mathias RT, FitzGerald PG, 2011. Intermediate filaments regulate tissue size and stiffness in the murine lens. Invest Ophthalmol Vis Sci 52, 3860–3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto M, Izu H, Seki K, Fukuda K, Nishida T, Yamada S, Kato K, Yonemura S, Inouye S, Nakai A, 2004. HSF4 is required for normal cell growth and differentiation during mouse lens development. The EMBO journal 23, 4297–4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhin DS, Nowak RB, Kim NE, Arnett EE, Chen AC, Sah RL, Clark JI, Fowler VM, 2012. Tmod1 and CP49 synergize to control the fiber cell geometry, transparency, and mechanical stiffness of the mouse lens. PloS one 7, e48734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong X, Cheng C, Xia CH, 2007. Connexins in lens development and cataractogenesis. The Journal of membrane biology 218, 9–12. [DOI] [PubMed] [Google Scholar]
- Gong X, Li E, Klier G, Huang Q, Wu Y, Lei H, Kumar NM, Horwitz J, Gilula NB, 1997. Disruption of alpha3 connexin gene leads to proteolysis and cataractogenesis in mice. Cell 91, 833–843. [DOI] [PubMed] [Google Scholar]
- Gorin MB, Yancey SB, Cline J, Revel JP, Horwitz J, 1984. The major intrinsic protein (MIP) of the bovine lens fiber membrane: characterization and structure based on cDNA cloning. Cell 39, 49–59. [DOI] [PubMed] [Google Scholar]
- Graham C, Hodin J, Wistow G, 1996. A retinaldehyde dehydrogenase as a structural protein in a mammalian eye lens. Gene recruitment of eta-crystallin. The Journal of biological chemistry 271, 15623–15628. [DOI] [PubMed] [Google Scholar]
- Graw J, 2009. Mouse models of cataract. Journal of genetics 88, 469–486. [DOI] [PubMed] [Google Scholar]
- Greiling TM, Houck SA, Clark JI, 2009. The zebrafish lens proteome during development and aging. Mol Vis 15, 2313–2325. [PMC free article] [PubMed] [Google Scholar]
- Grey AC, Walker KL, Petrova RS, Han J, Wilmarth PA, David LL, Donaldson PJ, Schey KL, 2013. Verification and spatial localization of aquaporin-5 in the ocular lens. Exp Eye Res 108, 94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslbeck M, Vierling E, 2015. A first line of stress defense: small heat shock proteins and their function in protein homeostasis. Journal of molecular biology 427, 1537–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JM, Hartsock A, Clark BS, Napier HR, Link BA, Gross JM, 2012. Integrin alpha5/fibronectin1 and focal adhesion kinase are required for lens fiber morphogenesis in zebrafish. Mol Biol Cell 23, 4725–4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Sun Y, 2007. Ribosomal protein S27L is a direct p53 target that regulates apoptosis. Oncogene 26, 2707–2716. [DOI] [PubMed] [Google Scholar]
- Heinicke LA, Nabet B, Shen S, Jiang P, van Zalen S, Cieply B, Russell JE, Xing Y, Carstens RP, 2013. The RNA binding protein RBM38 (RNPC1) regulates splicing during late erythroid differentiation. PloS one 8, e78031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG, 2006. eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem Sci 31, 553–562. [DOI] [PubMed] [Google Scholar]
- Hoang TV, Kumar PKR, Sutharzan S, Tsonis PA, Liang C, Robinson ML, 2014. Comparative transcriptome analysis of epithelial and fiber cells in newborn mouse lenses with RNA sequencing. Molecular Vision 20, 27. [PMC free article] [PubMed] [Google Scholar]
- Horwitz J, 2003. Alpha-crystallin. Exp Eye Res 76, 145–153. [DOI] [PubMed] [Google Scholar]
- Hou H, Wang F, Zhang W, Wang D, Li X, Bartlam M, Yao X, Rao Z, 2011. Structure-functional analyses of CRHSP-24 plasticity and dynamics in oxidative stress response. The Journal of biological chemistry 286, 9623–9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Rajagopal R, Liu Y, Dattilo LK, Shaham O, Ashery-Padan R, Beebe DC, 2011. The mechanism of lens placode formation: a case of matrix-mediated morphogenesis. Dev Biol 355, 32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireland ME, Wallace P, Sandilands A, Poosch M, Kasper M, Graw J, Liu A, Maisel H, Prescott AR, Hutcheson AM, Goebel D, Quinlan RA, 2000. Up-regulation of novel intermediate filament proteins in primary fiber cells: an indicator of all vertebrate lens fiber differentiation? The Anatomical record 258, 25–33. [DOI] [PubMed] [Google Scholar]
- Irum B, Khan SY, Ali M, Kaul H, Kabir F, Rauf B, Fatima F, Nadeem R, Khan AO, Al Obaisi S, Naeem MA, Nasir IA, Khan SN, Husnain T, Riazuddin S, Akram J, Eghrari AO, Riazuddin SA, 2016. Mutation in LIM2 Is Responsible for Autosomal Recessive Congenital Cataracts. PloS one 11, e0162620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV, 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11, 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing Z, Gangalum RK, Bhat AM, Nagaoka Y, Jiang M, Bhat SP, 2014. HSF4 mutation p.Arg116His found in age-related cataracts and in normal populations produces childhood lamellar cataract in transgenic mice. Human mutation 35, 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelberman D, Islam L, Holder SE, Jacques TS, Calvas P, Hennekam RC, Nischal KK, Sowden JC, 2011. Digenic inheritance of mutations in FOXC1 and PITX2 : correlating transcription factor function and Axenfeld-Rieger disease severity. Human mutation 32, 1144–1152. [DOI] [PubMed] [Google Scholar]
- Khan SY, Ali M, Kabir F, Renuse S, Na CH, Talbot CC Jr., Hackett SF, Riazuddin SA, 2018. Proteome Profiling of Developing Murine Lens Through Mass Spectrometry. Invest Ophthalmol Vis Sci 59, 100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan SY, Hackett SF, Lee MC, Pourmand N, Talbot CC Jr., Riazuddin SA, 2015. Transcriptome Profiling of Developing Murine Lens Through RNA Sequencing. Invest Ophthalmol Vis Sci 56, 4919–4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Lee JJ, Cho JH, Jeong J, Park AY, Kang W, Lee KJ, 2017. Heterogeneous nuclear ribonucleoprotein K inhibits heat shock-induced transcriptional activity of heat shock factor 1. The Journal of biological chemistry 292, 12801–12812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler J, Gilbert K, Brooks HV, Jolly RD, Hopcroft DH, Bullivant S, 1986. Membrane interlocking domains in the lens. Invest Ophthalmol Vis Sci 27, 1527–1534. [PubMed] [Google Scholar]
- Klein BE, Klein R, Lee KE, 1998. Incidence of age-related cataract: the Beaver Dam Eye Study. Arch Ophthalmol 116, 219–225. [DOI] [PubMed] [Google Scholar]
- Krey JF, Wilmarth PA, Shin JB, Klimek J, Sherman NE, Jeffery ED, Choi D, David LL, Barr-Gillespie PG, 2014. Accurate label-free protein quantitation with high- and low-resolution mass spectrometers. Journal of proteome research 13, 1034–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari S, Gao J, Mathias RT, Sun X, Eswaramoorthy A, Browne N, Zhang N, Varadaraj K, 2017. Aquaporin 0 Modulates Lens Gap Junctions in the Presence of Lens-Specific Beaded Filament Proteins. Invest Ophthalmol Vis Sci 58, 6006–6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuszak J, Alcala J, Maisel H, 1980. The surface morphology of embryonic and adult chick lens-fiber cells. The American journal of anatomy 159, 395–410. [DOI] [PubMed] [Google Scholar]
- Kuwabara T, 1975. The maturation of the lens cell: a morphologic study. Exp Eye Res 20, 427–443. [DOI] [PubMed] [Google Scholar]
- Lachke SA, Alkuraya FS, Kneeland SC, Ohn T, Aboukhalil A, Howell GR, Saadi I, Cavallesco R, Yue Y, Tsai AC, Nair KS, Cosma MI, Smith RS, Hodges E, Alfadhli SM, Al-Hajeri A, Shamseldin HE, Behbehani A, Hannon GJ, Bulyk ML, Drack AV, Anderson PJ, John SW, Maas RL, 2011. Mutations in the RNA granule component TDRD7 cause cataract and glaucoma. Science 331, 1571–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limi S, Senecal A, Coleman RA, Lopez-Jones M, Guo P, Polumbo C, Singer RH, Skoultchi AI, Cvekl A, 2018. Transcriptional burst fraction and size dynamics during lens fiber cell differentiation and detailed insights into the denucleation process. The Journal of biological chemistry [DOI] [PMC free article] [PubMed]
- Lionnet T, Czaplinski K, Darzacq X, Shav-Tal Y, Wells AL, Chao JA, Park HY, de Turris V, Lopez-Jones M, Singer RH, 2011. A transgenic mouse for in vivo detection of endogenous labeled mRNA. Nat Meth 8, 165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Lyu L, Chin D, Gao J, Sun X, Shang F, Caceres A, Chang ML, Rowan S, Peng J, Mathias R, Kasahara H, Jiang S, Taylor A, 2015. Altered ubiquitin causes perturbed calcium homeostasis, hyperactivation of calpain, dysregulated differentiation, and cataract. Proceedings of the National Academy of Sciences of the United States of America 112, 1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Kawai K, Khashabi S, Deng C, Liu YH, Yiu S, 2010. Inactivation of Smad4 leads to impaired ocular development and cataract formation. Biochemical and biophysical research communications 400, 476–482. [DOI] [PubMed] [Google Scholar]
- Lo WK, Harding CV, 1984. Square arrays and their role in ridge formation in human lens fibers. Journal of ultrastructure research 86, 228–245. [DOI] [PubMed] [Google Scholar]
- Logan CM, Bowen CJ, Menko AS, 2018. Functional role for stable microtubules in lens fiber cell elongation. Experimental cell research 362, 477–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou X, Kim J, Hawk BJ, Shin YK, 2017. alpha-Synuclein may cross-bridge v-SNARE and acidic phospholipids to facilitate SNARE-dependent vesicle docking. The Biochemical journal 474, 2039–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA, 2015. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161, 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddala R, Chauhan BK, Walker C, Zheng Y, Robinson ML, Lang RA, Rao PV, 2011a. Rac1 GTPase-deficient mouse lens exhibits defects in shape, suture formation, fiber cell migration and survival. Dev Biol 360, 30–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddala R, Skiba NP, Lalane R 3rd, Sherman DL, Brophy PJ, Rao PV, 2011b. Periaxin is required for hexagonal geometry and membrane organization of mature lens fibers. Dev Biol 357, 179–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher GJ, Black GC, Manson FD, 2012. Focus on molecules: lens intrinsic membrane protein (LIM2/MP20). Exp Eye Res 103, 115–116. [DOI] [PubMed] [Google Scholar]
- Mansergh FC, Hunter SM, Geatrell JC, Jarrin M, Powell K, Evans MJ, Wride MA, 2008. Developmentally regulated expression of hemoglobin subunits in avascular tissues. Int J Dev Biol 52, 873–886. [DOI] [PubMed] [Google Scholar]
- Mathias RT, White TW, Gong X, 2010. Lens gap junctions in growth, differentiation, and homeostasis. Physiological reviews 90, 179–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Martinez O, Brownell I, Amaya-Manzanares F, Hu Q, Behringer RR, Jamrich M, 2005. Severe defects in proliferation and differentiation of lens cells in Foxe3 null mice. Molecular and cellular biology 25, 8854–8863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael R, Bron AJ, 2011. The ageing lens and cataract: a model of normal and pathological ageing. Philos Trans R Soc Lond B Biol Sci 366, 1278–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- More MI, Kirsch FP, Rathjen FG, 2001. Targeted ablation of NrCAM or ankyrin-B results in disorganized lens fibers leading to cataract formation. J Cell Biol 154, 187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita H, Mizushima N, 2016. Autophagy in the lens. Exp Eye Res 144, 22–28. [DOI] [PubMed] [Google Scholar]
- Mulders SM, Preston GM, Deen PM, Guggino WB, van Os CH, Agre P, 1995. Water channel properties of major intrinsic protein of lens. The Journal of biological chemistry 270, 9010–9016. [DOI] [PubMed] [Google Scholar]
- Murphy P, Kabir MH, Srivastava T, Mason ME, Dewi CU, Lim S, Yang A, Djordjevic D, Killingsworth MC, Ho JWK, Harman DG, O’Connor MD, 2018. Light-focusing human micro-lenses generated from pluripotent stem cells model lens development and drug-induced cataract in vitro. Development 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag S, Larsson M, Robinson RC, Burtnick LD, 2013. Gelsolin: the tail of a molecular gymnast. Cytoskeleton (Hoboken, N.J.) 70, 360–384. [DOI] [PubMed] [Google Scholar]
- Nowak RB, Fischer RS, Zoltoski RK, Kuszak JR, Fowler VM, 2009. Tropomodulin1 is required for membrane skeleton organization and hexagonal geometry of fiber cells in the mouse lens. J Cell Biol 186, 915–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden AT, Nunes I, Ko K, Wu S, Hines CS, Wang AF, Hegde RS, Lang RA, 1998. GRIFIN, a novel lens-specific protein related to the galectin family. The Journal of biological chemistry 273, 28889–28896. [DOI] [PubMed] [Google Scholar]
- Ostrowska Z, Moraczewska J, 2017. Cofilin - a protein controlling dynamics of actin filaments. Postepy higieny i medycyny doswiadczalnej (Online) 71, 339–351. [DOI] [PubMed] [Google Scholar]
- Paterson CA, Delamere NA, 2004. ATPases and lens ion balance. Exp Eye Res 78, 699–703. [DOI] [PubMed] [Google Scholar]
- Perng MD, Zhang Q, Quinlan RA, 2007. Insights into the beaded filament of the eye lens. Experimental cell research 313, 2180–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrash JM, 2013. Aging and age-related diseases of the ocular lens and vitreous body. Invest Ophthalmol Vis Sci 54, Orsf54–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova RS, Schey KL, Donaldson PJ, Grey AC, 2015. Spatial distributions of AQP5 and AQP0 in embryonic and postnatal mouse lens development. Exp Eye Res 132, 124–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer JR, McAvoy BL, Fecteau RE, Deleault KM, Brooks SA, 2011. CARHSP1 is required for effective tumor necrosis factor alpha mRNA stabilization and localizes to processing bodies and exosomes. Molecular and cellular biology 31, 277–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plageman TF Jr., Chung MI, Lou M, Smith AN, Hildebrand JD, Wallingford JB, Lang RA, 2010. Pax6-dependent Shroom3 expression regulates apical constriction during lens placode invagination. Development 137, 405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posner M, Murray KL, McDonald MS, Eighinger H, Andrew B, Drossman A, Haley Z, Nussbaum J, David LL, Lampi KJ, 2017. The zebrafish as a model system for analyzing mammalian and native alpha-crystallin promoter function. PeerJ 5, e4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts JD, Kornacker S, Beebe DC, 1998. Activation of the Jak-STAT-signaling pathway in embryonic lens cells. Dev Biol 204, 277–292. [DOI] [PubMed] [Google Scholar]
- Quinlan RA, 2015. DRUG DISCOVERY. A new dawn for cataracts. Science 350, 636–637. [DOI] [PubMed] [Google Scholar]
- Rao PV, Maddala R, 2006. The role of the lens actin cytoskeleton in fiber cell elongation and differentiation. Semin Cell Dev Biol 17, 698–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redman KL, Rechsteiner M, 1989. Identification of the long ubiquitin extension as ribosomal protein S27a. Nature 338, 438–440. [DOI] [PubMed] [Google Scholar]
- Robinson ML, Overbeek PA, 1996. Differential expression of alpha A- and alpha B-crystallin during murine ocular development. Invest Ophthalmol Vis Sci 37, 2276–2284. [PubMed] [Google Scholar]
- Rong P, Wang X, Niesman I, Wu Y, Benedetti LE, Dunia I, Levy E, Gong X, 2002. Disruption of Gja8 (alpha8 connexin) in mice leads to microphthalmia associated with retardation of lens growth and lens fiber maturation. Development 129, 167–174. [DOI] [PubMed] [Google Scholar]
- Rozenblatt-Rosen O, Stubbington MJT, Regev A, Teichmann SA, 2017. The Human Cell Atlas: from vision to reality. Nature 550, 451–453. [DOI] [PubMed] [Google Scholar]
- Sandilands A, Prescott AR, Carter JM, Hutcheson AM, Quinlan RA, Richards J, FitzGerald PG, 1995a. Vimentin and CP49/filensin form distinct networks in the lens which are independently modulated during lens fibre cell differentiation. Journal of cell science 108 ( Pt 4), 1397–1406. [DOI] [PubMed] [Google Scholar]
- Sandilands A, Prescott AR, Hutcheson AM, Quinlan RA, Casselman JT, FitzGerald PG, 1995b. Filensin is proteolytically processed during lens fiber cell differentiation by multiple independent pathways. European journal of cell biology 67, 238–253. [PubMed] [Google Scholar]
- Schey KL, Petrova RS, Gletten RB, Donaldson PJ, 2017. The Role of Aquaporins in Ocular Lens Homeostasis. International journal of molecular sciences 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schey KL, Wang Z, J LW, Qi Y, 2014. Aquaporins in the eye: expression, function, and roles in ocular disease. Biochimica et biophysica acta 1840, 1513–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt N, Kollewe A, Constantin CE, Henrich S, Ritzau-Jost A, Bildl W, Saalbach A, Hallermann S, Kulik A, Fakler B, Schulte U, 2017. Neuroplastin and Basigin Are Essential Auxiliary Subunits of Plasma Membrane Ca(2+)-ATPases and Key Regulators of Ca(2+) Clearance. Neuron 96, 827–838.e829. [DOI] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M, 2011. Global quantification of mammalian gene expression control. Nature 473, 337–342. [DOI] [PubMed] [Google Scholar]
- Shang F, Wilmarth PA, Chang ML, Liu K, David LL, Caceres MA, Wawrousek E, Taylor A, 2014. Newborn mouse lens proteome and its alteration by lysine 6 mutant ubiquitin. Journal of proteome research 13, 1177–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma KK, Santhoshkumar P, 2009. Lens aging: effects of crystallins. Biochimica et biophysica acta 1790, 1095–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, De Maria AB, Wang H, Mathias RT, FitzGerald PG, Bassnett S, 2011. Further analysis of the lens phenotype in Lim2-deficient mice. Invest Ophthalmol Vis Sci 52, 7332–7339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiels A, Hejtmancik JF, 2017. Mutations and mechanisms in congenital and age-related cataracts. Exp Eye Res 156, 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiels A, King JM, Mackay DS, Bassnett S, 2007. Refractive defects and cataracts in mice lacking lens intrinsic membrane protein-2. Invest Ophthalmol Vis Sci 48, 500–508. [DOI] [PubMed] [Google Scholar]
- Shurtleff MJ, Temoche-Diaz MM, Karfilis KV, Ri S, Schekman R, 2016. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. eLife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddam AD, Gautier-Courteille C, Perez-Campos L, Anand D, Kakrana A, Dang CA, Legagneux V, Mereau A, Viet J, Gross JM, Paillard L, Lachke SA, 2018. The RNA-binding protein Celf1 post-transcriptionally regulates p27Kip1 and Dnase2b to control fiber cell nuclear degradation in lens development. PLoS Genet 14, e1007278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simirskii VN, Lee RS, Wawrousek EF, Duncan MK, 2006. Inbred FVB/N mice are mutant at the cp49/Bfsp2 locus and lack beaded filament proteins in the lens. Invest Ophthalmol Vis Sci 47, 4931–4934. [DOI] [PubMed] [Google Scholar]
- Sindhu Kumari S, Gupta N, Shiels A, FitzGerald PG, Menon AG, Mathias RT, Varadaraj K, 2015. Role of Aquaporin 0 in lens biomechanics. Biochemical and biophysical research communications 462, 339–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E, 2005. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science 310, 1513–1515. [DOI] [PubMed] [Google Scholar]
- Skruber K, Read TA, Vitriol EA, 2018. Reconsidering an active role for G-actin in cytoskeletal regulation. Journal of cell science 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slingsby C, Wistow GJ, Clark AR, 2013. Evolution of crystallins for a role in the vertebrate eye lens. Protein science : a publication of the Protein Society 22, 367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder E, Soundararajan R, Sharma M, Dearth A, Smith B, Braun RE, 2015. Compound Heterozygosity for Y Box Proteins Causes Sterility Due to Loss of Translational Repression. PLoS Genet 11, e1005690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Landsbury A, Dahm R, Liu Y, Zhang Q, Quinlan RA, 2009. Functions of the intermediate filament cytoskeleton in the eye lens. The Journal of clinical investigation 119, 1837–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava R, Budak G, Dash S, Lachke SA, Janga SC, 2017. Transcriptome analysis of developing lens reveals abundance of novel transcripts and extensive splicing alterations. Sci Rep 7, 11572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupfler B, Birck C, Seraphin B, Mauxion F, 2016. BTG2 bridges PABPC1 RNA-binding domains and CAF1 deadenylase to control cell proliferation. Nat Commun 7, 10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Rockowitz S, Chauss D, Wang P, Kantorow M, Zheng D, Cvekl A, 2015a. Chromatin features, RNA polymerase II and the comparative expression of lens genes encoding crystallins, transcription factors, and autophagy mediators. Mol Vis 21, 955–973. [PMC free article] [PubMed] [Google Scholar]
- Sun J, Rockowitz S, Xie Q, Ashery-Padan R, Zheng D, Cvekl A, 2015b. Identification of in vivo DNA-binding mechanisms of Pax6 and reconstruction of Pax6-dependent gene regulatory networks during forebrain and lens development. Nucleic acids research 43, 6827–6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Zhao Y, McGreal R, Cohen-Tayar Y, Rockowitz S, Wilczek C, Ashery-Padan R, Shechter D, Zheng D, Cvekl A, 2016. Pax6 associates with H3K4-specific histone methyltransferases Mll1, Mll2, and Set1a and regulates H3K4 methylation at promoters and enhancers. Epigenetics Chromatin 9, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup A, Bell BA, Du J, Han JYS, Soto J, Abel ED, Bravo-Nuevo A, FitzGerald PG, Peachey NS, Philp NJ, 2018. Deletion of GLUT1 in mouse lens epithelium leads to cataract formation. Exp Eye Res [DOI] [PMC free article] [PubMed]
- Thanos S, Bohm MR, Meyer zu Horste M, Prokosch-Willing V, Hennig M, Bauer D, Heiligenhaus A, 2014. Role of crystallins in ocular neuroprotection and axonal regeneration. Prog Retin Eye Res 42, 145–161. [DOI] [PubMed] [Google Scholar]
- Toyama BH, Hetzer MW, 2013. Protein homeostasis: live long, won’t prosper. Nat Rev Mol Cell Biol 14, 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truscott RJ, Friedrich MG, 2016. The etiology of human age-related cataract. Proteins don’t last forever. Biochimica et biophysica acta 1860, 192–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Raamsdonk CD, Tilghman SM, 2000. Dosage requirement and allelic expression of PAX6 during lens placode formation. Development 127, 5439–5448. [DOI] [PubMed] [Google Scholar]
- van Rheede T, Amons R, Stewart N, de Jong WW, 2003. Lactate dehydrogenase A as a highly abundant eye lens protein in platypus (Ornithorhynchus anatinus): upsilon (upsilon)-crystallin. Molecular biology and evolution 20, 994–998. [DOI] [PubMed] [Google Scholar]
- van Zalen S, Lombardi AA, Jeschke GR, Hexner EO, Russell JE, 2015. AUF-1 and YB-1 independently regulate beta-globin mRNA in developing erythroid cells through interactions with poly(A)-binding protein. Mech Dev 136, 40–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaino JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, Xu QW, Wang R, Hermjakob H, 2016. 2016 update of the PRIDE database and its related tools. Nucleic acids research 44, D447–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A, Regev A, Yosef N, 2016. Revealing the vectors of cellular identity with single-cell genomics. Nature biotechnology 34, 1145–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J, Menko AS, 2009. Integrins in lens development and disease. Exp Eye Res 88, 216–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Hom G, Zhou S, Guo M, Li B, Yang J, Monnier VM, Fan X, 2017. The oxidized thiol proteome in aging and cataractous mouse and human lens revealed by ICAT labeling. Aging Cell 16, 244–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E, Geng A, Maniar AM, Mui BW, Gong X, 2016. Connexin 50 Regulates Surface Ball-and-Socket Structures and Fiber Cell Organization. Invest Ophthalmol Vis Sci 57, 3039–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Han J, David LL, Schey KL, 2013. Proteomics and phosphoproteomics analysis of human lens fiber cell membranes. Invest Ophthalmol Vis Sci 54, 1135–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Schey KL, 2009. Phosphorylation and truncation sites of bovine lens connexin 46 and connexin 50. Exp Eye Res 89, 898–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Schey KL, 2015. Proteomic Analysis of Lipid Raft-Like Detergent-Resistant Membranes of Lens Fiber Cells. Invest Ophthalmol Vis Sci 56, 8349–8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenke JL, McDonald WH, Schey KL, 2016. Spatially Directed Proteomics of the Human Lens Outer Cortex Reveals an Intermediate Filament Switch Associated With the Remodeling Zone. Invest Ophthalmol Vis Sci 57, 4108–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West-Mays JA, Pino G, Lovicu FJ, 2010. Development and use of the lens epithelial explant system to study lens differentiation and cataractogenesis. Prog Retin Eye Res 29, 135–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitson JA, Wilmarth PA, Klimek J, Monnier VM, David L, Fan X, 2017. Proteomic analysis of the glutathione-deficient LEGSKO mouse lens reveals activation of EMT signaling, loss of lens specific markers, and changes in stress response proteins. Free radical biology & medicine 113, 84–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willekens B, Vrensen G, 1981. The three-dimensional organization of lens fibers in the rabbit. A scanning electron microscopic reinvestigation. Albrecht von Graefes Archiv fur klinische und experimentelle Ophthalmologie. Albrecht von Graefe’s archive for clinical and experimental ophthalmology 216, 275–289. [DOI] [PubMed] [Google Scholar]
- Willekens B, Vrensen G, 1985. Lens fiber organization in four avian species: a scanning electron microscopic study. Tissue & cell 17, 359–377. [DOI] [PubMed] [Google Scholar]
- Wilmarth PA, Riviere MA, David LL, 2009. Techniques for accurate protein identification in shotgun proteomic studies of human, mouse, bovine, and chicken lenses. Journal of ocular biology, diseases, and informatics 2, 223–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wistow G, Bernstein SL, Wyatt MK, Behal A, Touchman JW, Bouffard G, Smith D, Peterson K, 2002. Expressed sequence tag analysis of adult human lens for the NEIBank Project: over 2000 non-redundant transcripts, novel genes and splice variants. Mol Vis 8, 171–184. [PubMed] [Google Scholar]
- Wistow GJ, Lietman T, Williams LA, Stapel SO, de Jong WW, Horwitz J, Piatigorsky J, 1988. Tau-crystallin/alpha-enolase: one gene encodes both an enzyme and a lens structural protein. J Cell Biol 107, 2729–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf L, Harrison W, Huang J, Xie Q, Xiao N, Sun J, Kong L, Lachke SA, Kuracha MR, Govindarajan V, Brindle PK, Ashery-Padan R, Beebe DC, Overbeek PA, Cvekl A, 2013. Histone posttranslational modifications and cell fate determination: lens induction requires the lysine acetyltransferases CBP and p300. Nucleic acids research 41, 10199–10214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo MK, Lee A, Fischer RS, Moyer J, Fowler VM, 2000. The lens membrane skeleton contains structures preferentially enriched in spectrin-actin or tropomodulin-actin complexes. Cell motility and the cytoskeleton 46, 257–268. [DOI] [PubMed] [Google Scholar]
- Wormstone IM, Eldred JA, 2016. Experimental models for posterior capsule opacification research. Exp Eye Res 142, 2–12. [DOI] [PubMed] [Google Scholar]
- Xie Q, Cvekl A, 2011. The orchestration of mammalian tissue morphogenesis through a series of coherent feed-forward loops. The Journal of biological chemistry 286, 43259–43271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Yang Y, Brennan L, Bouhassira EE, Kantorow M, Cvekl A, 2010. Efficient generation of lens progenitor cells and lentoid bodies from human embryonic stem cells in chemically defined conditions. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 24, 3274–3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Stopka T, Golestaneh N, Wang Y, Wu K, Li A, Chauhan BK, Gao CY, Cveklova K, Duncan MK, Pestell RG, Chepelinsky AB, Skoultchi AI, Cvekl A, 2006. Regulation of alphaA-crystallin via Pax6, c-Maf, CREB and a broad domain of lens-specific chromatin. The EMBO journal 25, 2107–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon KH, Blankenship T, Shibata B, Fitzgerald PG, 2008. Resisting the effects of aging: a function for the fiber cell beaded filament. Invest Ophthalmol Vis Sci 49, 1030–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihama M, Uechi T, Asakawa S, Kawasaki K, Kato S, Higa S, Maeda N, Minoshima S, Tanaka T, Shimizu N, Kenmochi N, 2002. The human ribosomal protein genes: sequencing and comparative analysis of 73 genes. Genome Res 12, 379–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HM, Liu T, Liu CJ, Song S, Zhang X, Liu W, Jia H, Xue Y, Guo AY, 2015. AnimalTFDB 2.0: a resource for expression, prediction and functional study of animal transcription factors. Nucleic acids research 43, D76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Hawse J, Huang Q, Sheets N, Miller KM, Horwitz J, Kantorow M, 2002. Decreased expression of ribosomal proteins in human age-related cataract. Invest Ophthalmol Vis Sci 43, 198–204. [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Zheng D, Cvekl A, 2018. A comprehensive spatial-temporal transcriptomic analysis of differentiating nascent mouse lens epithelial and fiber cells. Exp Eye Res 175, 56–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


