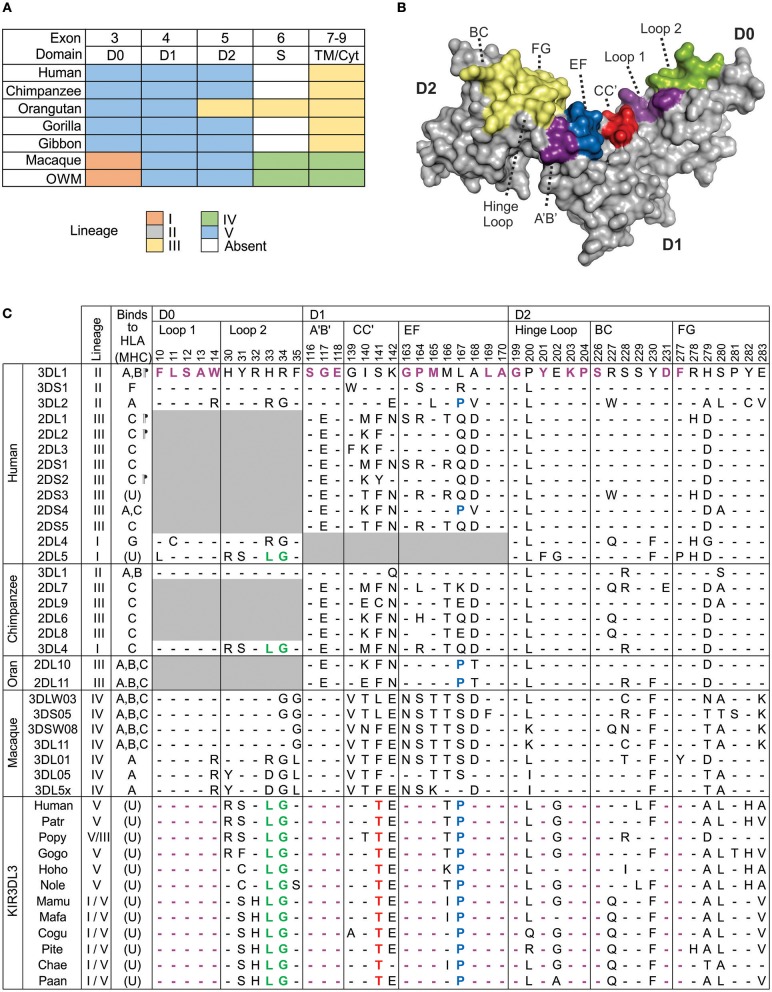
Figure 1.
Conserved Ig-like domains of catarrhine KIR3DL3. (A) Shown is a summary of the phylogenetic relationships of KIR3DL3 sequences when compared with other KIR sequences from catarrhine primates. Each domain was analyzed separately and colored boxes correspond to the distinct KIR lineages; lineages are given in the color key at the bottom. Exon 6, which encodes the stem is absent from all ape KIR3DL3 except orangutan. Lineage IV is monkey specific (30) and may be part of lineage II (16). OWM—old world monkeys other than macaque. (B) Shows a predictive model of the Ig-domains of KIR3DL3. The surface regions corresponding to the HLA class I binding loops of KIR3DL1 are shaded; those containing residues conserved in KIR3DL3 are shaded violet, green, blue, or red to correspond with (C), and others are shaded in yellow. (C) Shown are the ligand binding motifs of KIR, as characterized by crystallography. At the left is shown the ancestral KIR lineage, and any known HLA/MHC ligand (A, B, C, F, or G), or unknown (U). (¶) indicates determined by crystal structure (45–48). Top: Residues are numbered according to the mature KIR3DL1 protein. In the alignment underneath, only the amino acid differences from KIR3DL1, which is used as reference, are shown. (-) indicates identity with the reference. Gray boxes indicate domains or residues that are absent from the respective allotype. Below the human KIR, are the respective binding-site motifs from chimpanzee, orangutan, and macaque KIR that are known to bind MHC class I (32, 49–52). The lowest box shows the equivalent motifs from human, other ape, and old-world monkey KIR3DL3; amino acids conserved across all KIR3DL3 molecules are colored red (unique to KIR3DL3), or violet, green, blue (identical to other KIR known to bind MHC, as indicated). Patr, chimpanzee; Popy, orangutan; Gogo, gorilla; Hoho/Nole, gibbon; Mamu, macaque; Pite, red colobus monkey; Cogu, black and white colobus monkey; Chae, African green monkey; Paan, olive baboon.