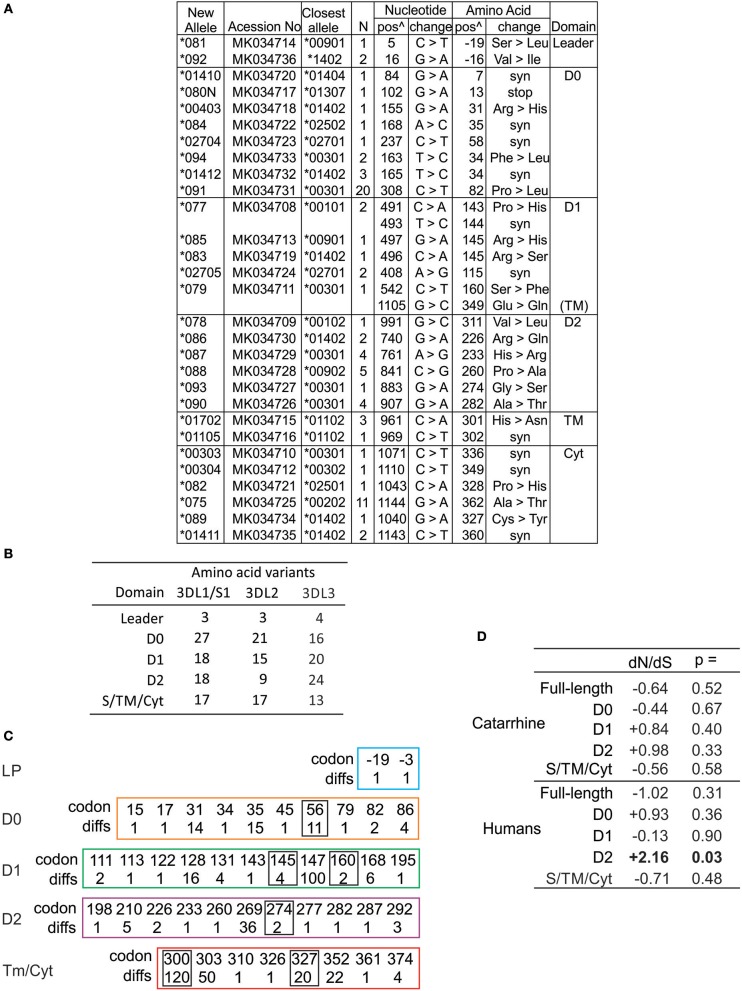
Figure 3.
High diversity of human KIR3DL3 Ig domains. (A) Shown are the novel KIR3DL3 alleles characterized for this study from Europeans and Papua New Guineans. From left to right: GenBank ID, the closest matched of the previously established alleles, number of individuals with new allele, nucleotide changes compared to closest match, corresponding amino acid (AA) substitutions, and the domains where the latter occur (Leader–leader peptide, D0-D2–Ig-like domains, TM–transmembrane, Cyt–cytoplasmic). Nucleotides are numbered according to the full length CDS, and amino acids according to the mature protein. (syn) indicates synonymous nucleotide substitution. Additional SNP variants characterized from 1000 Genomes individuals are given in Figure S4. (B) Shown is the number of amino acid variants in each of the domains of human KIR3DL1/S1, KIR3DL2, and KIR3DL3. (C) Shown are the codon numbers for every amino acid that is polymorphic in KIR3DL3 (codon) and the number of alleles that differ at that position (diffs). Boxes indicate residues having more than one alternative amino acid. The domains are depicted separately. (D) Shown are standard dN/dS analyses of KIR3DL3 nucleotide sequences. Results consistent with (-) negative (purifying) selection or (+) positive (diversifying) selection are indicated, and those statistically significant indicated in bold text. Analysis of catarrhine primate sequences is shown at the top and humans only at the bottom. In each case both the full length CDS and individual domains were analyzed.