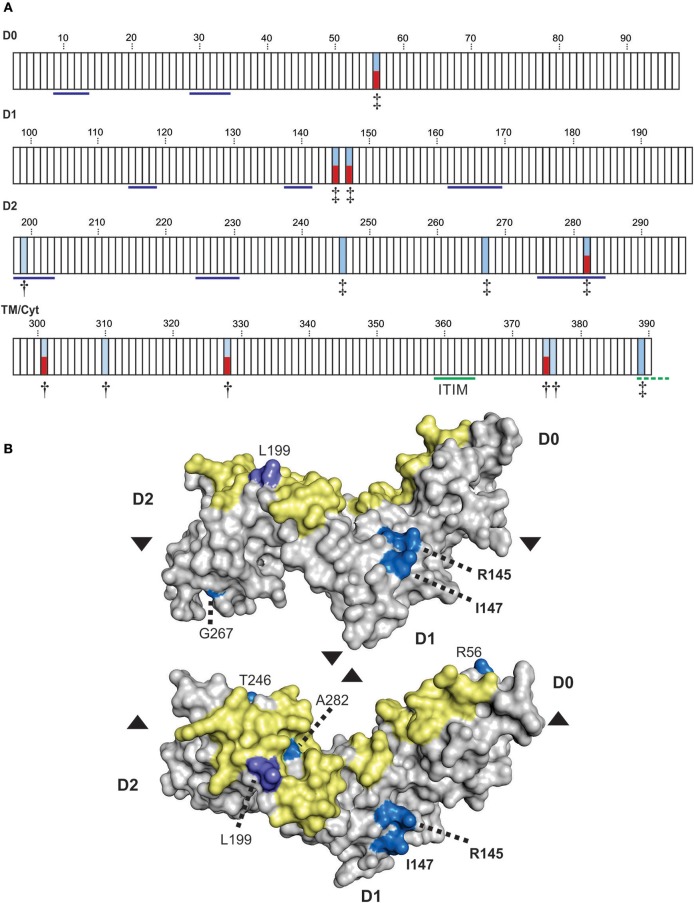
Figure 5.
Positive selection diversifies residues of KIR3DL3 involved in protein aggregation. (A) Shown are the results from codon-by-codon selection analysis of KIR3DL3. Each box corresponds to an amino acid residue of human KIR3DL3, numbered according to the mature protein. Blue shading corresponds to positively selected sites (†P > 0.9, ‡P > 0.95). Red shading indicates those positively selected sites that are also polymorphic in humans. Residues underlined in blue in D0-D2 correspond to the HLA class I binding sites of KIR3DL1 (45). The functional immunoreceptor tyrosine-based inhibitory motif (ITIM) is underlined in green, the non-functional second ITIM, which is interrupted by a stop codon in human KIR3DL3, is underlined in dashed green. The evidence for selection in each domain is supported by a posterior probability of >0.90. The four domains (D0, D1, D2, and TM/Cyt) were analyzed separately. (B) A space-filling model of the Ig-domains of KIR3DL3. Positively selected residues are denoted by shades of blue- bright blue: PP > 0.95, light purple: PP > 0.90. The region corresponding to the HLA class I binding site of KIR3DL1 is shaded in yellow. The amino acid residues are numbered as (A). Upper–“side” view, lower–“top” view; where arrows indicate the relative rotation.