Figure 5.
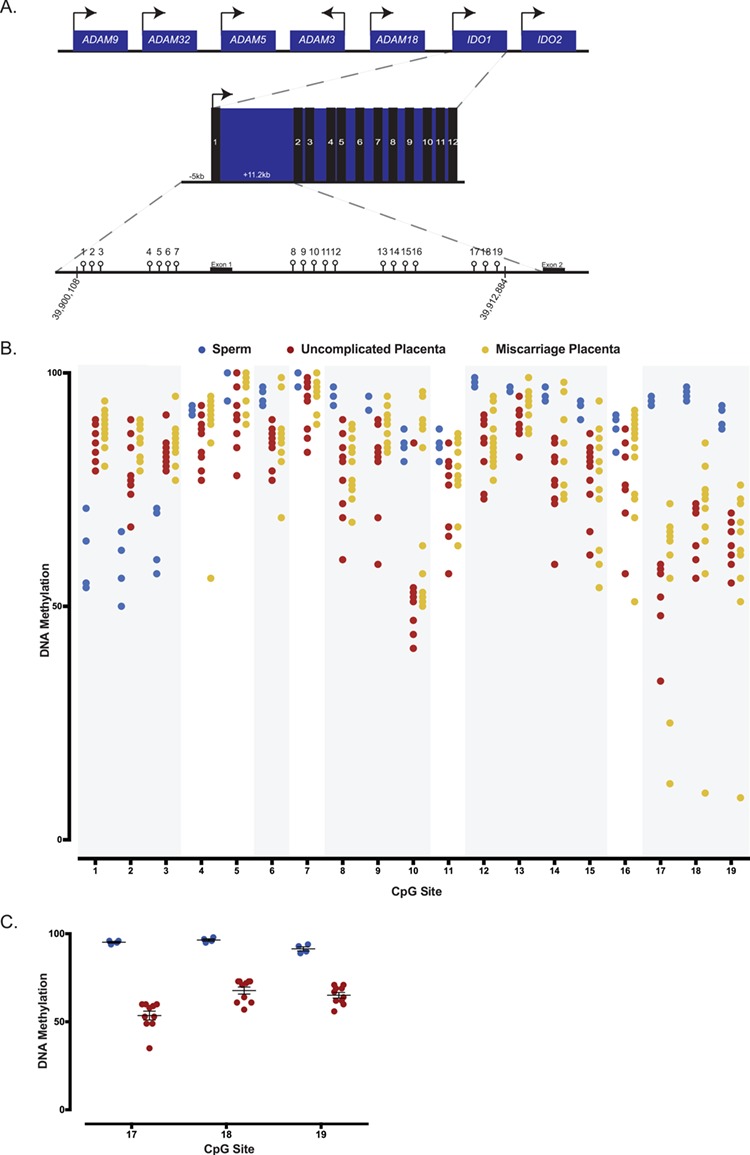
DNA methylation analysis of human sperm and first trimester placentas from uncomplicated and pregnancy loss using bisulfite pyrosequencing. (A) Schematic representation of the human IDO1 locus on chromosome 8 containing 12 exons, and of the 19 CpG sites assayed within the human IDO1 locus. Arrows designate direction of gene expression. (B) Average DNA methylation levels for human sperm, uncomplicated first trimester placentas and euploid first trimester miscarriage placentas for all 19 CpG sites. A total of four sperm samples, 10 uncomplicated placentas and 13 miscarriage placentas are analyzed and presented. Global analysis of average CpG sites reveals significant differences in methylation between sperm and uncomplicated placentas. Individual CpG sites analysis shows that CpG sites 1–3, 6, 8–10, 12–15 and 17–19 significantly differed between sperm and uncomplicated placentas (regions shaded in grey). Comparing uncomplicated to miscarriage placentas, average methylation significantly differ when the CpG sites are analyzed globally. (C) Average DNA methylation of CpG sites 17, 18 and 19 in human sperm (blue) and placentas from uncomplicated pregnancies (red). Each data point represents an individual. CpG sites represent a candidate imprinted DMR as they are highly methylated in sperm but partially methylated in the placentas. Differences in percent or variance in methylation for individual CpGs were assessed using a t-test or homogeneity test of variance, and the corresponding P-values were adjusted for multiple comparisons using the Benjamini–Hochberg method. P-values and FDR of <0.05 were considered to be statistically significant.
