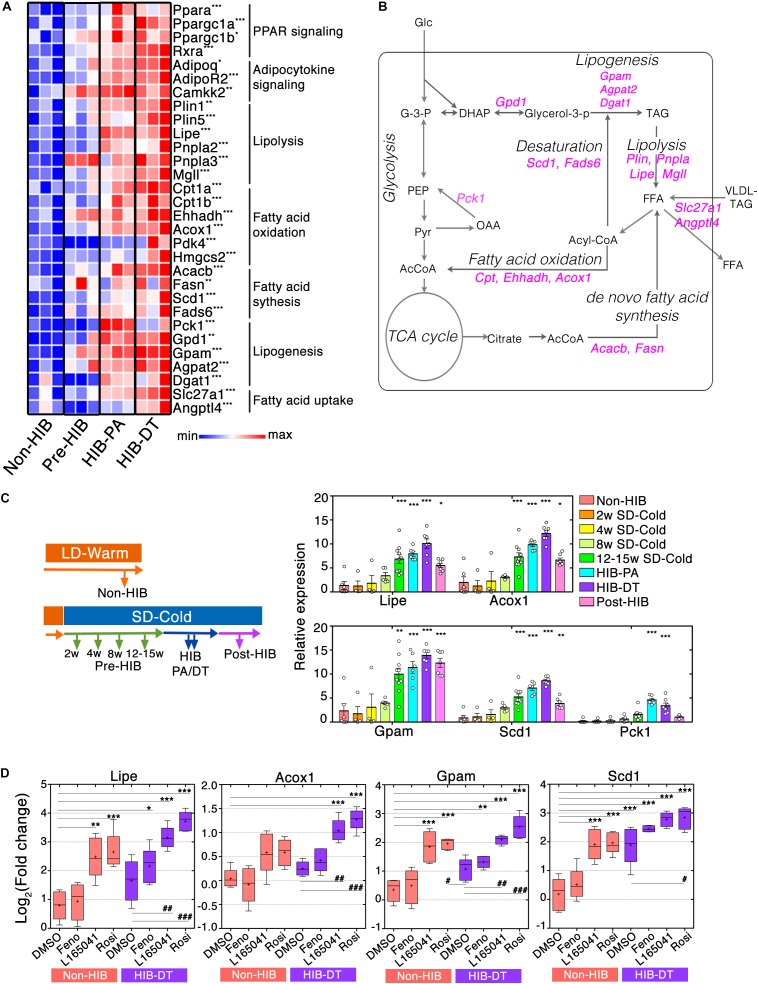
FIGURE 4.
PPAR signaling augments mRNA expression of genes required for lipid catabolism and anabolism in iWAT during hibernation. (A) Heat maps of fold changes in mRNA expression of genes related to PPAR signaling pathways. ∗q < 0.05, ∗∗q < 0.01, ∗∗∗q < 0.005. (B) A schematic representation of the cellular biochemical networks of lipid and glucose metabolism in WAT. Genes that encode enzymes for chemical reaction and that are up-regulated during HIB are shown in magenta. AcCoA (acetyl-CoA), DHAP (dihydroxyacetone phosphate), FFA (free fatty acid), G-3-P (glyceraldehyde 3-phosphate), Glc (glucose), Glycerol-3-P (glycerol 3-phosphate), OAA (oxaloacetate), PEP (phosphoenolpyruvate), Pyr (pyruvate), TAG (triacylglyceride), VLDL-TAG (very-low-density lipoprotein triacylglyceride). (C) Time-course analysis of the lipid metabolism genes that were up-regulated during HIB. qPCR analysis of relative mRNA expression levels of lipid metabolism genes at the different time points. The results are expressed as mean ± SEM. White circles show each animal. P-value was derived from one-way ANOVA and Tukey’s multiple comparison tests. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005 against the “Non-HIB group.” (D) Ex vivo experiments of adipose explant cultures. iWAT explants from Syrian hamsters were treated with 0.1% dimethyl sulfoxide (DMSO), 100 μM fenofibrate (Feno), 100 μM L165041 (L165041), and 100 μM rosiglitazone (Rosi) for 48 h. Relative mRNA expression level of the lipid metabolism genes in the different conditions was determined using qPCR. Horizontal lines and crosses indicate medians and means of relative mRNA expression, respectively. Boxes enclose the interquartile ranges and whiskers show the minimum and maximum relative mRNA expression of each experimental condition. P-value was derived using one-way ANOVA and Tukey’s multiple comparison tests. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005 against the DMSO-treated condition of the Non-HIB group. #p < 0.05, ##p < 0.01, ###p < 0.005 against the DMSO-treated condition of HIB-DT group.